Viomycin
 | |
| Clinical data | |
|---|---|
| Routes of administration | Intramuscular injection |
| ATC code |
|
| Identifiers | |
| |
| CAS Number | |
| PubChem CID | |
| ChemSpider | |
| UNII | |
| ChEBI | |
| ChEMBL | |
| ECHA InfoCard | 100.046.643 |
| Chemical and physical data | |
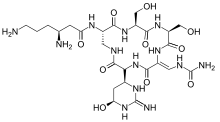
| Formula | C25H43N13O10 |
| Molar mass | 685.691 g/mol |
| 3D model (JSmol) | |
| |
| |
| | |
Viomycin is a member of the tuberactinomycin family,[1][2][3] a group of nonribosomal peptide antibiotics exhibiting anti-tuberculosis properties. The tuberactinomycin family is an essential component in the drug cocktail currently used to fight infections of Mycobacterium tuberculosis. Viomycin was the first member of the tuberactinomycins to be isolated and identified[4] and was used to treat TB until it was replaced by the less toxic, but structurally related compound, capreomycin. The tuberactinomycins target bacterial ribosomes, binding RNA and disrupting bacterial protein biosynthesis. It is produced by the actinomycete Streptomyces puniceus, that binds to RNA and inhibits prokaryotic protein synthesis and certain forms of RNA splicing.
Biosynthesis
The gene cluster for viomycin has been sequenced from Streptomyces sp. strain ATCC 11861,[5] Streptomyces vinaceus[6] and from Streptomyces lividans 1326.[4] It consists of a central cyclic pentapeptide code assembled from nonribosomal peptide synthetase (NRPS). The NRPS contains 4 proteins: VioA, VioF, VioI, and VioG. These proteins condense and cyclize two molecules of L-2,3-diaminopropionate (L-Dap), two molecules of L-serine (L-Ser), and one molecule of (2S,3R)-capreomycidine (L-Cam). After cyclizing these, VioJ catalyzes the α,β-desaturation of this preliminary structure. It is proposed that the viomycin gene cluster includes 36.3 kb of contiguous DNA that encodes 20 open reading frames (ORFs)[5] that are involved in the biosynthesis, regulation, and eventual activation viomycin. In addition to these ORFs, the structure contains the resistance gene vph. The following is a summary of the ORFs and their functions.
- VioA: NRPS (A-PCP-C-A-PCP-C)
- VioH: Type II thioesterase
- VioO: NRPS (A-PCP)-β-lysine activation
- VioB: 2,3-diaminopropionate synthase
- VioI: NRPS (PCP-C)
- VioP: Lysine 2,3-aminomutase
- VioC: L-Arg hydroxylase
- VIoJ: 2,3-diaminopropionyl α,β-desaturase
- vph: Viomycin phosphotransferase
- VioD: Capreomycidine synthase
- VioK: Ornithine cyclodeaminase
- VioQ: Capreomycidine hydroxylase
- VioE: Permease
- VioL: Carbamoyltransferase
- VioR: Transcriptional regulator
- VioF: NRPS (A-PCP-C)
- VIoM: NRPS (C)-β-lysine transferase
- VIoS: Viomycin-phosphate phosphatase
- VioG: NRPS (A-PCP-C/)
- VioN: MbtH homolog
- VioT: Transcriptional regulator
Synthesis of the backbone
The following is the proposed biosynthesis of viomycin using NRPS-catalyzed peptide synthesis. There are five modules for cyclic pentapeptide biosynthesis, including one that lacks an adenylation domain (A). It is therefore proposed that one of the other A domains functions twice. Additionally, the NRPS subunits are not suspected to function in the order in which their genes are arranged, a characteristic of viomycin biosynthesis that is unlike typical NRPS-catalyzed peptide synthesis.[4] The NRPS components function in the order of VioA→VioI→VioF→VioG to account for the incorporation of β-ureidoalanine (β-Uda). The first A domain of VioA creates an L-Dap-PCP intermediate on the first PCP domain. Meanwhile, the second A domain of VioA loads L-Ser onto the second PCP domain, as well as the PCP of VioI. The activation of β-Uda occurs via VioF, and VioG incorporates L-Cam.
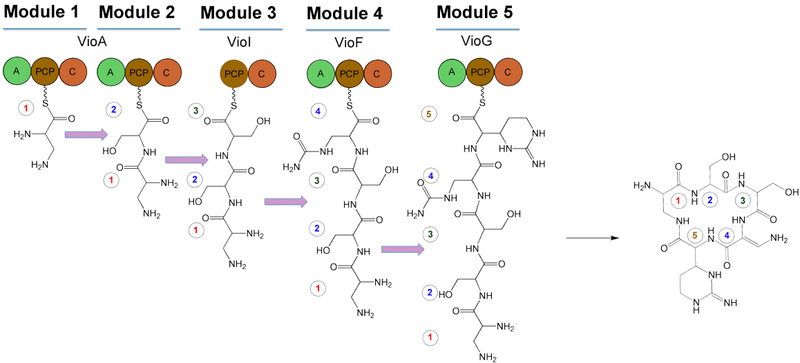
Post-modification
After α,β-desaturation via VioJ, three modifications to the preliminary cyclic structure occur. Hydroxylation of C-6 in the structure occurs by VioQ, N-acylation of α-amino group using β-lysine, VioO, and VioM, and carbamoylation of the β-amino group, producing β-ureidoalanine (β-Uda) by the carbamoyltransferase homologue VioL.

References
- ↑ B. W. Bycroft (1972). "The crystal structure of viomycin, a tuberculostatic antibiotic". Chem. Commun.: 660. doi:10.1039/c39720000660.
- ↑ Noda; Take, T; Nagata, A; Wakamiya, T; Shiba, T; et al. (1972). "Chemical studies on tuberactinomycin. 3. The chemical structure of viomycin (tuberactinomycin B)". J. Antibiot. 25 (7): 427–8. PMID 4350196. doi:10.7164/antibiotics.25.427.
- ↑ T. Kitagawa; et al. (1972). "The total structure of viomycin by sequential analysis". Chem. Pharm. Bull. 20 (10): 2215–25. PMID 4346588. doi:10.1248/cpb.20.2215.
- 1 2 3 Barkei, J.; Kevany, B.; Felnagle, E.; Thomas, M. (2009). "Investigations into viomycin biosynthesis by using heterologous production in Streptomyces lividans.". ChemBioChem. 10 (2): 366–76. PMC 2765823
 . PMID 19105177. doi:10.1002/cbic.200800646.
. PMID 19105177. doi:10.1002/cbic.200800646. - 1 2 Thomas, M.; Chan, Y.; Ozanick, S. (2003). "Deciphering tuberactinomycin biosynthesis: isolation, sequencing, and annotation of the viomycin biosynthetic gene cluster.". Antimicrob. Agents Chemother. 47 (9): 2823–30. PMC 182626
 . PMID 12936980. doi:10.1128/AAC.47.9.2823-2830.2003.
. PMID 12936980. doi:10.1128/AAC.47.9.2823-2830.2003. - ↑ Yin, X.; O'Hare, T.; Gould, S.; Zabriskie, M. (2003). "Identification and cloning of genes encoding viomycin biosynthesis from Streptomyces vinaceus and evidence for involvement of a rare oxygenase.". Gene. 312: 215–24. PMID 12909358. doi:10.1016/S0378-1119(03)00617-6.