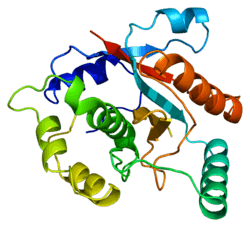
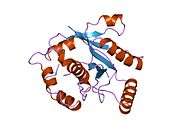
UCHL3
Ubiquitin carboxyl-terminal hydrolase isozyme L3 is an enzyme that in humans is encoded by the UCHL3 gene.[5][6]
Interactions
UCHL3 has been shown to interact with NEDD8 and the tauopathy and synucleinopathy associated mutated ubiquitin molecule UBB+1.[7][8]
See also
- Ubiquitin carboxy-terminal hydrolase L1—an enzyme
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000118939 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000022111 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J (Dec 1989). "The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase". Science. 246 (4930): 670–3. PMID 2530630. doi:10.1126/science.2530630.
- ↑ "Entrez Gene: UCHL3 ubiquitin carboxyl-terminal esterase L3 (ubiquitin thiolesterase)".
- ↑ Dennissen FJ, Kholod N, Hermes DJ, Kemmerling N, Steinbusch HW, Dantuma NP, van Leeuwen FW (July 2011). "Mutant ubiquitin (UBB(+1)) associated with neurodegenerative disorders is hydrolyzed by ubiquitin C-terminal hydrolase L3 (UCH-L3)". FEBS Lett. 585 (16): 2568–74. PMID 21762696. doi:10.1016/j.febslet.2011.06.037.
- ↑ Wada H, Kito K, Caskey LS, Yeh ET, Kamitani T (October 1998). "Cleavage of the C-terminus of NEDD8 by UCH-L3". Biochem. Biophys. Res. Commun. 251 (3): 688–92. PMID 9790970. doi:10.1006/bbrc.1998.9532.
Further reading
- Johnston SC, Larsen CN, Cook WJ, et al. (1997). "Crystal structure of a deubiquitinating enzyme (human UCH-L3) at 1.8 A resolution.". EMBO J. 16 (13): 3787–96. PMC 1170002
 . PMID 9233788. doi:10.1093/emboj/16.13.3787.
. PMID 9233788. doi:10.1093/emboj/16.13.3787. - Wilkinson KD, Laleli-Sahin E, Urbauer J, et al. (1999). "The binding site for UCH-L3 on ubiquitin: mutagenesis and NMR studies on the complex between ubiquitin and UCH-L3.". J. Mol. Biol. 291 (5): 1067–77. PMID 10518943. doi:10.1006/jmbi.1999.3038.
- Baek SH, Yoo YJ, Tanaka K, Chung CH (1999). "Molecular cloning of chick UCH-6 which shares high similarity with human UCH-L3: its unusual substrate specificity and tissue distribution.". Biochem. Biophys. Res. Commun. 264 (1): 235–40. PMID 10527871. doi:10.1006/bbrc.1999.1492.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences.". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. PMC 139241
 . PMID 12477932. doi:10.1073/pnas.242603899.
. PMID 12477932. doi:10.1073/pnas.242603899. - Saito S, Iida A, Sekine A, et al. (2003). "Catalog of 680 variations among eight cytochrome p450 ( CYP) genes, nine esterase genes, and two other genes in the Japanese population.". J. Hum. Genet. 48 (5): 249–70. PMID 12721789. doi:10.1007/s10038-003-0021-7.
- Nam MJ, Madoz-Gurpide J, Wang H, et al. (2004). "Molecular profiling of the immune response in colon cancer using protein microarrays: occurrence of autoantibodies to ubiquitin C-terminal hydrolase L3.". Proteomics. 3 (11): 2108–15. PMID 14595809. doi:10.1002/pmic.200300594.
- Dunham A, Matthews LH, Burton J, et al. (2004). "The DNA sequence and analysis of human chromosome 13.". Nature. 428 (6982): 522–8. PMC 2665288
 . PMID 15057823. doi:10.1038/nature02379.
. PMID 15057823. doi:10.1038/nature02379. - Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC).". Genome Res. 14 (10B): 2121–7. PMC 528928
 . PMID 15489334. doi:10.1101/gr.2596504.
. PMID 15489334. doi:10.1101/gr.2596504. - Misaghi S, Galardy PJ, Meester WJ, et al. (2005). "Structure of the ubiquitin hydrolase UCH-L3 complexed with a suicide substrate.". J. Biol. Chem. 280 (2): 1512–20. PMID 15531586. doi:10.1074/jbc.M410770200.
- Rolén U, Kobzeva V, Gasparjan N, et al. (2006). "Activity profiling of deubiquitinating enzymes in cervical carcinoma biopsies and cell lines.". Mol. Carcinog. 45 (4): 260–9. PMID 16402389. doi:10.1002/mc.20177.
- Lim J, Hao T, Shaw C, et al. (2006). "A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration.". Cell. 125 (4): 801–14. PMID 16713569. doi:10.1016/j.cell.2006.03.032.
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.