U4 spliceosomal RNA
| U4 spliceosomal RNA | |
|---|---|
 Predicted secondary structure and sequence conservation of U4 | |
| Identifiers | |
| Symbol | U4 |
| Rfam | RF00015 |
| Other data | |
| RNA type | Gene; snRNA; splicing |
| Domain(s) | Eukaryota |
| GO | 0017070 0000353 0000351 0005687 0046540 |
| SO | 0000393 |

The U4 small nuclear Ribo-Nucleic Acid (U4 snRNA) is a non-coding RNA component of the major or U2-dependent spliceosome – a eukaryotic molecular machine involved in the splicing of pre-messenger RNA (pre-mRNA). It forms a duplex with U6, and with each splicing round, it is displaced from the U6 snRNA (and the spliceosome) in an ATP-dependent manner, allowing U6 to re-fold and create the active site for splicing catalysis. A recycling process involving protein Brr2 releases U4 from U6, while protein Prp24 re-anneals U4 and U6. The crystal structure of a 5′ stem-loop of U4 in complex with a binding protein has been solved.[1]
Biological role
The U4 snRNA has been shown to exist in a number of different formats including: bound to proteins as a small nuclear Ribo-Nuclear Protein snRNP,[2] involved with the U6 snRNA in the di-snRNP,[3] as well as involved with both the U6 snRNA and the U5 snRNA in the tri-snRNP.[4][5] The different formats have been proposed to coincide with different temporal events in the activity of the penta-snRNP,[6] or as intermediates in the step-wise model of spliceosome assembly and activity.[7]
The U4 snRNA (and its likely analog snR14 in Yeast[8]) has been shown not to participate directly in the specific catalytic activities of the splicing reaction,[9] and is proposed instead to act as a regulator of the U6 snRNA. The U4 snRNA inhibits spliceosome activity during assembly by complementary base pairing between the U6 snRNA in two highly conserved stem regions.[10] It is suggested that this base-pairing interaction prevents the U6 snRNA from assembling with the U2 snRNA into the conformation required for catalytic activity.[11] If the U4 snRNA is degraded and thereby removed from the spliceosome, splicing is effectively halted.[12] The U4 and U6 snRNAs are demonstratively required for splicing in vitro.[13]
Structure
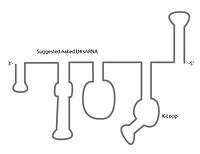

The U4 snRNA secondary structure is suggested to alter depending on its interaction with the U6 snRNA.[7] Several experiments involving X-ray crystallography,[1][14] NMR,[15] and chemical modification RNA structure probing[16] indicate that U4 snRNA secondary structure contains several conserved motifs,[17] which serve structural as well as intermediary roles in establishing interactions with other splicing components. The putative U4/U6 snRNA base pairing secondary structure shown in Figure 2., is conserved across a diverse set of organisms suggesting the splicing machinery's ancient origins.[18] It has been shown previously that a highly conserved Kinked-loop participates in specific protein interactions.[1][19]
Interactions
The U4 snRNA must be displaced from U6 snRNA in an ATP dependent process involving the protein Brr2 - before the spliceosome is made active.[9][20][21] A cycle has been proposed including both Brr2 as well as the protein prp24 which selectively re-anneals U4 to the U6 snRNA.[21][22][23][24] A ring of Sm proteins surround a conserved region of the U4 snRNA near the 3' end which are expected to promote favorable interactions between the different snRNPs as well as possibly protect the U4 snRNA from degradation by RNAse enzymes.[25][26] Over 100 proteins have been identified that participate in spliceosomal pathway, several proteins of varying size are also known to interact with the U4 snRNP.[27]
References
- 1 2 3 4 Vidovic I, Nottrott S, Hartmuth K, Lührmann R, Ficner R (December 2000). "Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment". Mol. Cell. 6 (6): 1331–42. PMID 11163207. doi:10.1016/S1097-2765(00)00131-3.
- ↑ Raghunathan PL, Guthrie C (July 1998). "RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2". Curr. Biol. 8 (15): 847–55. PMID 9705931. doi:10.1016/S0960-9822(07)00345-4.
- ↑ Bringmann P, Appel B, Rinke J, Reuter R, Theissen H, Lührmann R (June 1984). "Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing". EMBO J. 3 (6): 1357–63. PMC 557523
 . PMID 6204860.
. PMID 6204860. - ↑ Black DL, Pinto AL (August 1989). "U5 small nuclear ribonucleoprotein: RNA structure analysis and ATP-dependent interaction with U4/U6". Mol. Cell. Biol. 9 (8): 3350–9. PMC 362380
 . PMID 2552294.
. PMID 2552294. - ↑ Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J (November 2001). "Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae". RNA. 7 (11): 1543–53. PMC 1370197
 . PMID 11720284.
. PMID 11720284. - ↑ Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J (January 2002). "Composition and functional characterization of the yeast spliceosomal penta-snRNP". Mol. Cell. 9 (1): 31–44. PMID 11804584. doi:10.1016/S1097-2765(02)00436-7.
- 1 2 Cheng SC, Abelson J (November 1987). "Spliceosome assembly in yeast". Genes Dev. 1 (9): 1014–27. PMID 2962902. doi:10.1101/gad.1.9.1014.
- ↑ Siliciano PG, Brow DA, Roiha H, Guthrie C (August 1987). "An essential snRNA from S. cerevisiae has properties predicted for U4, including interaction with a U6-like snRNA". Cell. 50 (4): 585–92. PMID 2440583. doi:10.1016/0092-8674(87)90031-6.
- 1 2 Yean SL, Lin RJ (November 1991). "U4 small nuclear RNA dissociates from a yeast spliceosome and does not participate in the subsequent splicing reaction". Mol. Cell. Biol. 11 (11): 5571–7. PMC 361927
 . PMID 1833635.
. PMID 1833635. - ↑ Guthrie C, Patterson B (1988). "Spliceosomal snRNAs". Annu. Rev. Genet. 22: 387–419. PMID 2977088. doi:10.1146/annurev.ge.22.120188.002131.
- ↑ Madhani HD, Guthrie C (November 1992). "A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome". Cell. 71 (5): 803–17. PMID 1423631. doi:10.1016/0092-8674(92)90556-R.
- ↑ Berget SM, Robberson BL (August 1986). "U1, U2, and U4/U6 small nuclear ribonucleoproteins are required for in vitro splicing but not polyadenylation". Cell. 46 (5): 691–6. PMID 2427201. doi:10.1016/0092-8674(86)90344-2.
- ↑ Black DL, Steitz JA (August 1986). "Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein". Cell. 46 (5): 697–704. PMID 2427202. doi:10.1016/0092-8674(86)90345-4.
- ↑ Kambach C, Walke S, Nagai K (April 1999). "Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles". Curr. Opin. Struct. Biol. 9 (2): 222–30. PMID 10322216. doi:10.1016/S0959-440X(99)80032-3.
- ↑ Comolli LR, Ulyanov NB, Soto AM, Marky LA, James TL, Gmeiner WH (October 2002). "NMR structure of the 3' stem-loop from human U4 snRNA". Nucleic Acids Res. 30 (20): 4371–9. PMC 137124
 . PMID 12384583. doi:10.1093/nar/gkf560.
. PMID 12384583. doi:10.1093/nar/gkf560. - ↑ Mougin A, Gottschalk A, Fabrizio P, Lührmann R, Branlant C (April 2002). "Direct probing of RNA structure and RNA-protein interactions in purified HeLa cell's and yeast spliceosomal U4/U6.U5 tri-snRNP particles". J. Mol. Biol. 317 (5): 631–49. PMID 11955014. doi:10.1006/jmbi.2002.5451.
- ↑ Li L, Otake LR, Xu Y, Michaeli S (January 2000). "The trans-spliceosomal U4 RNA from the monogenetic trypanosomatid Leptomonas collosoma. Cloning and identification of a transcribed trna-like element that controls its expression". J. Biol. Chem. 275 (4): 2259–64. PMID 10644672. doi:10.1074/jbc.275.4.2259.
- ↑ Izquierdo JM, Valcárcel J (July 2006). "A simple principle to explain the evolution of pre-mRNA splicing". Genes Dev. 20 (13): 1679–84. PMID 16818600. doi:10.1101/gad.1449106.
- ↑ Boon KL, Norman CM, Grainger RJ, Newman AJ, Beggs JD (February 2006). "Prp8p dissection reveals domain structure and protein interaction sites". RNA. 12 (2): 198–205. PMC 1370899
 . PMID 16373487. doi:10.1261/rna.2281306.
. PMID 16373487. doi:10.1261/rna.2281306. - ↑ Blencowe BJ, Sproat BS, Ryder U, Barabino S, Lamond AI (November 1989). "Antisense probing of the human U4/U6 snRNP with biotinylated 2'-OMe RNA oligonucleotides". Cell. 59 (3): 531–9. PMID 2478298. doi:10.1016/0092-8674(89)90036-6.
- 1 2 Raghunathan PL, Guthrie C (February 1998). "A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles". Science. 279 (5352): 857–60. PMID 9452384. doi:10.1126/science.279.5352.857.
- ↑ Fortner DM, Troy RG, Brow DA (January 1994). "A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing". Genes Dev. 8 (2): 221–33. PMID 8299941. doi:10.1101/gad.8.2.221.
- ↑ Jandrositz A, Guthrie C (February 1995). "Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs". EMBO J. 14 (4): 820–32. PMC 398149
 . PMID 7882985.
. PMID 7882985. - ↑ Ghetti A, Company M, Abelson J (April 1995). "Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs". RNA. 1 (2): 132–45. PMC 1369067
 . PMID 7585243.
. PMID 7585243. - ↑ Urlaub H, Raker VA, Kostka S, Lührmann R (January 2001). "Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure". EMBO J. 20 (1–2): 187–96. PMC 140196
 . PMID 11226169. doi:10.1093/emboj/20.1.187.
. PMID 11226169. doi:10.1093/emboj/20.1.187. - ↑ Stark H, Dube P, Lührmann R, Kastner B (January 2001). "Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle". Nature. 409 (6819): 539–42. PMID 11206553. doi:10.1038/35054102.
- ↑ Nottrott S, Urlaub H, Lührmann R (October 2002). "Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins". EMBO J. 21 (20): 5527–38. PMC 129076
 . PMID 12374753. doi:10.1093/emboj/cdf544.
. PMID 12374753. doi:10.1093/emboj/cdf544.
Further reading
- Zwieb, C (1997). "The uRNA database". Nucleic Acids Res. 25 (1): 102–103. PMC 146409
 . PMID 9016512. doi:10.1093/nar/25.1.102.
. PMID 9016512. doi:10.1093/nar/25.1.102. - Thomas, J; Lea K; Zucker-Aprison E; Blumenthal T (1990). "The spliceosomal snRNAs of Caenorhabditis elegans". Nucleic Acids Res. 18 (9): 2633–2642. PMC 330746
 . PMID 2339054. doi:10.1093/nar/18.9.2633.
. PMID 2339054. doi:10.1093/nar/18.9.2633.