Tetrahydrocannabinolic acid synthase
| Tetrahydrocannibinolic acid synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
_synthase_protein_structure.png) 3VTE | |||||||||
| Identifiers | |||||||||
| EC number | 1.21.3.7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Tetrahydrocannabinolic acid (THCA) synthase (full name Δ1-tetrahydrocannabinolic acid synthase) is an enzyme responsible for catalyzing the formation of THCA from cannabigerolic acid (CBGA). THCA is the direct precursor of tetrahydrocannabinol (THC), the principal psychoactive component of cannabis, which is produced from various strains of Cannibis sativa. Therefore, THCA synthase is considered to be a key enzyme controlling cannabis psychoactivity.[1] Polymorphisms of THCA synthase result in varying levels of THC in Cannabis plants, resulting in "drug-type" and "fiber-type" C. sativa varieties.[2][3]
Structure
THCA synthase is a 60 kDa (~500 amino acids) monomeric enzyme with the isoelectric point at 6.4.[4] Post-translational N-linked glycosylation increases the total mass to approximately 74 kDa. The tertiary structure is divided into two domains (domains I and II), with a flavin adenine dinucleotide (FAD) positioned between the two domains. Domain I comprises eight alpha helices and eight beta sheet and is covalently bound to FAD. Domain II comprises five alpha helices surrounded by eight beta sheets. Enzymes that share similar amino acid sequences include the flavoproteins berberine bridge enzyme (BBE), glucooligosaccharide oxidase (GOOX), and aclacinomycin oxidoreductase (AknOx).[1]
The FAD moiety is the location of enzymatic activity and is covalently bound to His114 and Cys176. FAD is also bound by hydrogen bonds with neighboring amino acid main chains and side chains. The co-crystallization of THCA synthase with substrate or product has not been accomplished yet.[1]
 FAD cofactor (in green) is located between domain I and domain II of THCA synthase. Alpha helices are cyan and beta sheets are magenta.
FAD cofactor (in green) is located between domain I and domain II of THCA synthase. Alpha helices are cyan and beta sheets are magenta. FAD (in green) is covalently bound to histidine 114 and cysteine 176 (in pink).
FAD (in green) is covalently bound to histidine 114 and cysteine 176 (in pink).
Reaction mechanism

THCA synthase, a flavoprotein, uses a flavin adenine dinucleotide (FAD) cofactor to catalyze the oxidative cyclization of the monoterpene moiety of cannabigerolic acid (CBGA). Similar cyclization reactions occur in monoterpene biosynthesis from geranyl pyrophosphate, but are not oxidative.[5] THCA synthase exhibits no catalytic activity against cannabigerol, which lacks a carboxyl group compared to CBGA, suggesting that the carboxyl group of CPGA is necessary for the reaction to occur.[1]
The overall chemical reaction is: CPGA + O2 THCA + H2O2
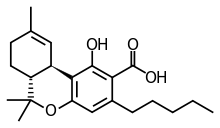
A hydride is transferred from CBGA to reduce FAD, concerted by deprotonation of a hydroxyl group by a tyrosine residue. The monoterpene moiety in CBGA is then positioned to complete cyclization into THCA. Oxidation of reduced FAD by O2 produces hydrogen peroxide (H2O2).[6][1]

Biological function
THCA synthase is expressed in the glandular trichomes of Cannabis sativa. THCA synthase may contribute to the self-defense of Cannabis plants by producing THCA and hydrogen peroxide, which are both cytotoxic. Because these products are toxic to the plant, THCA synthase is secreted into the trichome storage cavity.[7] THCA also acts as a necrosis-inducing factor by opening mitochondrial permeability transition pores, inhibiting mitochondrial viability and resulting in senescence in leaf tissues.[8]
Non-enzymatic decarboxylation of THCA during storage or smoking forms THC, the principal psychoactive component of cannabis.[9] Further degradation by temperature, auto-oxidation, and light forms cannabinol.[10] THC and other cannabinoids are well-known to reduce nausea and vomiting and stimulate hunger, particularly in patients undergoing cancer chemotherapy.[11]
Similar enzymes to THCA synthase catalyze the formation of other cannabinoids. For example, cannabidiolic acid (CBDA) synthase is a flavoprotein that catalyzes a similar oxidative cyclization of CPGA into CBDA, the dominant cannabinoid constituent of fiber-type C. sativa. CBDA undergoes a similar decarboxylation to form cannabidiol.[12]
Significance
Demand is high for pharmaceutical grade THC and other cannabinoids due to interest in their potential therapeutic use, but is stymied by legal regulations of C. sativa cultivation in many countries.[13] Direct chemical synthesis of THC is difficult due to high costs and low yields.[14] Therefore, the use of THCA synthase for the production of THC has been explored, as CBGA is easy is synthesize and THCA readily decarboxylates to form THC.[10] Biosynthesis of THCA by expressing THCA synthase in organisms has been attempted in bacteria, insects, and tobacco plants with limited success. Production of THCA on a milligram scale has been demonstrated in Pichia pastoris yeast cells in two independent studies.[15][13]
References
- 1 2 3 4 5 Shoyama, Yoshinari; Tamada, Taro; Kurihara, Kazuo; Takeuchi, Ayako; Taura, Futoshi; Arai, Shigeki; Blaber, Michael; Shoyama, Yukihiro; Morimoto, Satoshi (2012-10-12). "Structure and function of ∆1-tetrahydrocannabinolic acid (THCA) synthase, the enzyme controlling the psychoactivity of Cannabis sativa". Journal of Molecular Biology. 423 (1): 96–105. ISSN 1089-8638. PMID 22766313. doi:10.1016/j.jmb.2012.06.030.
- ↑ Staginnus, Christina; Zörntlein, Siegfried; de Meijer, Etienne (2014-07-01). "A PCR marker linked to a THCA synthase polymorphism is a reliable tool to discriminate potentially THC-rich plants of Cannabis sativa L". Journal of Forensic Sciences. 59 (4): 919–926. ISSN 1556-4029. PMID 24579739. doi:10.1111/1556-4029.12448.
- ↑ Kojoma, Mareshige; Seki, Hikaru; Yoshida, Shigeo; Muranaka, Toshiya. "DNA polymorphisms in the tetrahydrocannabinolic acid (THCA) synthase gene in "drug-type" and "fiber-type" Cannabis sativa L.". Forensic Science International. 159 (2–3): 132–140. doi:10.1016/j.forsciint.2005.07.005.
- ↑ Taura, F.; Morimoto, S.; Shoyama, Y.; Mechoulam, R. (1995). "First direct evidence for the mechanism of Δ1-tetrahydrocannabinolic acid biosynthesis". J. Am. Chem. Soc. 117: 9766–9767. doi:10.1021/ja00143a024.
- ↑ Sirikantaramas, Supaart; Morimoto, Satoshi; Shoyama, Yoshinari; Ishikawa, Yu; Wada, Yoshiko; Shoyama, Yukihiro; Taura, Futoshi (2004-09-17). "The Gene Controlling Marijuana Psychoactivity MOLECULAR CLONING AND HETEROLOGOUS EXPRESSION OF Δ1-TETRAHYDROCANNABINOLIC ACID SYNTHASE FROM CANNABIS SATIVA L.". Journal of Biological Chemistry. 279 (38): 39767–39774. ISSN 0021-9258. PMID 15190053. doi:10.1074/jbc.M403693200.
- ↑ Shoyama, Y.; Takeuchi, A.; Taura, F.; Tamada, T.; Adachi, M.; Kuroki, R.; Shoyama, Y.; Morimoto, S. (2005). "Crystallization of Δ1-tetrahydrocannabinolic acid (THCA) synthase from Cannabis sativa". Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61 (Pt 8): 799–801. PMC 1952348
 . PMID 16511162. doi:10.1107/S1744309105023365.
. PMID 16511162. doi:10.1107/S1744309105023365. - ↑ Sirikantaramas, S. (2005-09-01). "Tetrahydrocannabinolic Acid Synthase, the Enzyme Controlling Marijuana Psychoactivity, is Secreted into the Storage Cavity of the Glandular Trichomes". Plant and Cell Physiology. 46 (9): 1578–1582. ISSN 0032-0781. doi:10.1093/pcp/pci166.
- ↑ Morimoto, Satoshi; Tanaka, Yumi; Sasaki, Kaori; Tanaka, Hiroyuki; Fukamizu, Tomohide; Shoyama, Yoshinari; Shoyama, Yukihiro; Taura, Futoshi (2007-07-13). "Identification and Characterization of Cannabinoids That Induce Cell Death through Mitochondrial Permeability Transition in Cannabis Leaf Cells". Journal of Biological Chemistry. 282 (28): 20739–20751. ISSN 0021-9258. PMID 17513301. doi:10.1074/jbc.M700133200.
- ↑ Yamauchi, T.; Shoyama, Y.; Aramaki, H.; Azuma, T.; Nishioka, I. (1967-07-01). "Tetrahydrocannabinolic acid, a genuine substance of tetrahydrocannabinol". Chemical & Pharmaceutical Bulletin. 15 (7): 1075–1076. ISSN 0009-2363. PMID 5583149. doi:10.1248/cpb.15.1075.
- 1 2 Moreno-Sanz, Guillermo (2016-06-01). "Can You Pass the Acid Test? Critical Review and Novel Therapeutic Perspectives of Δ9-Tetrahydrocannabinolic Acid A". Cannabis and Cannabinoid Research. 1 (1): 124–130. doi:10.1089/can.2016.0008.
- ↑ Guzmán, Manuel (2003-10-01). "Cannabinoids: potential anticancer agents". Nature Reviews Cancer. 3 (10): 745–755. ISSN 1474-175X. doi:10.1038/nrc1188.
- ↑ Taura, Futoshi; Sirikantaramas, Supaart; Shoyama, Yoshinari; Yoshikai, Kazuyoshi; Shoyama, Yukihiro; Morimoto, Satoshi (2007-06-26). "Cannabidiolic-acid synthase, the chemotype-determining enzyme in the fiber-type Cannabis sativa". FEBS Letters. 581 (16): 2929–2934. ISSN 1873-3468. PMID 17544411. doi:10.1016/j.febslet.2007.05.043.
- 1 2 Zirpel, Bastian; Stehle, Felix; Kayser, Oliver (2015-09-01). "Production of Δ9-tetrahydrocannabinolic acid from cannabigerolic acid by whole cells of Pichia (Komagataella) pastoris expressing Δ9-tetrahydrocannabinolic acid synthase from Cannabis sativa L". Biotechnology Letters. 37 (9): 1869–1875. ISSN 1573-6776. PMID 25994576. doi:10.1007/s10529-015-1853-x.
- ↑ Trost, Barry M.; Dogra, Kalindi (2007-03-01). "Synthesis of (-)-Δ9-trans-Tetrahydrocannabinol: Stereocontrol via Mo-Catalyzed Asymmetric Allylic Alkylation Reaction". Organic Letters. 9 (5): 861–863. ISSN 1523-7060. PMC 2597621
 . PMID 17266321. doi:10.1021/ol063022k.
. PMID 17266321. doi:10.1021/ol063022k. - ↑ Lange, Kerstin; Schmid, Andreas; Julsing, Mattijs K. (2015-10-10). "Δ(9)-Tetrahydrocannabinolic acid synthase production in Pichia pastoris enables chemical synthesis of cannabinoids". Journal of Biotechnology. 211: 68–76. ISSN 1873-4863. PMID 26197418. doi:10.1016/j.jbiotec.2015.06.425.