Recombination signal sequences
Recombination signal sequences are conserved sequences of noncoding DNA that are recognized by the RAG1/RAG2 enzyme complex during V(D)J recombination in immature B cells and T cells.[1] Recombination signal sequences guide the enzyme complex to the V,D, and J gene segments that will undergo recombination during the formation of the heavy and light-chain variable regions in T-cell receptors and immunoglobulin molecules.[1]
Structure

RSSs are made up of conserved heptamer sequences (7 base pairs), spacer sequences, and conserved nonamer sequences (9 base pairs) that are adjacent to the V, D and J sequences in the heavy-chain region of DNA and the V and J sequences in the light-chain DNA region.[1][2] Spacer sequences are located between heptamer and nonamer sequences and exhibit base pair variety but are always either 12 base pairs or 23 base pairs long.[3] Unlike spacer sequences, heptamer sequences are usually CACAGTG, and the first three nucleotides are highly conserved.[3] Nonamers are usually ACAAAAACC, and the A/T basepairs are also highly conserved.[3] The RAG1/RAG2 enzyme complex follows the 12-23 rule when joining V,D, and J segments, pairing 12-bp spacer RSSs to 23-bp spacer RSSs.[1][2] This prevents two different genes coding for the same region from recombining (ex. V-V recombination).[1] RSSs are located between V,D, and J segments of the germ-line DNA of maturing B and T lymphocytes and are permanently spliced out of the final Ig mRNA product after V(D)J recombination is complete.[1]
Function
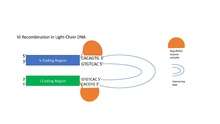
The RAG1/RAG2 enzyme complex recognizes the heptamer sequences flanking the V and J coding regions and nicks their 5' end, releasing the intervening DNA between the V and J coding regions.[1] In the heavy-chain coding region of DNA, the RAG1/RAG2 enzyme complex recognizes the RSSs flanking the D and J segments and brings them together, forming a loop containing intervening DNA.[1][4] The RAG1/RAG2 complex then introduces a nick at the 5' end of the RSS heptamers adjacent to the coding regions on both the D and J segments, permanently removing the loop of intervening DNA and creating a double-stranded break that is repaired by VDJ recombinase enzymes.[1][4] This process is repeated for the joining of V to DJ.[1] In light-chain rearrangement, only V and J segments are brought together.[1]
Related Diseases & Disorders
cRSS
Cryptic RSSs are gene sequences that resemble authentic RSSs and are occasionally mistaken for them by the RAG1/RAG2 enzyme complex.[3] Recombining an RSS with a cRSS can lead to chromosome translocations, which can lead to cancer.[3]
Omenn's Syndrome
Some infants born with autosomal recessive SCIDS lack a functional copies of the genes that code for the RAG1/RAG2 enzyme complex because of missense mutations.[5][6] These infants will produce a non-functional RAG1/RAG2 enzyme complex that cannot recognize RSSs and therefore cannot initiate V(D)J recombination effectively.[5][6] This disorder is characterized by a lack of functioning B and T cells.[1][5]
References
- 1 2 3 4 5 6 7 8 9 10 11 12 Owen, Judith A.; Punt, Jenni; Stranford, Sharon A. (2013). Kuby Immunology. New York: W.H. Freeman and Company. pp. 233–235.
- 1 2 Schatz, David G.; Oettinger, Marjorie A.; Schlissel, Mark S. (1992). "V(D)J Recombination: Molecular Biology and Regulation". Annual Review Immunology. 10: 259–283.
- 1 2 3 4 5 Roth, David B. (2014). "V(D)J Recombination: Mechanism, Errors, and Fidelity". Microbiology Spectrum: 1–11.
- 1 2 Rodgers, Karla (2017). "Riches in RAGs: Revealing the V(D)J Recombinase Through High-Resolution Structures". Cell Press. 42: 72–84.
- 1 2 3 Buckley, Rebecca H. (2004). "Molecular Defects in Human Severe Combined Immunodeficiency And Approaches to Immune Reconstitution". Annual Review Immunology: 625–655.
- 1 2 Elnour, Ibtisam B; Ahmed, Shakeel; Halim, Kamel; Nirmala, V (2007). "Omenn's Syndrome: A rare primary immunodeficiency disorder". Sultan Qaboos University Medical Journal: 1–6.