Porphobilinogen synthase
| porphobilinogen synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
DALA dehydratase | |||||||||
| Identifiers | |||||||||
| EC number | 4.2.1.24 | ||||||||
| CAS number | 9036-37-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
| Delta-aminolevulinic acid dehydratase | |
|---|---|
| Identifiers | |
| Symbol | ALAD |
| Entrez | 210 |
| HUGO | 395 |
| OMIM | 125270 |
| RefSeq | NM_001003945 |
| UniProt | P13716 |
| Other data | |
| EC number | 4.2.1.24 |
| Locus | Chr. 9 q32 |
| ALAD | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 high resolution crystal structure of a mg2-dependent 5-aminolevulinic acid dehydratase | |||||||||
| Identifiers | |||||||||
| Symbol | ALAD | ||||||||
| Pfam | PF00490 | ||||||||
| Pfam clan | CL0036 | ||||||||
| InterPro | IPR001731 | ||||||||
| PROSITE | PDOC00153 | ||||||||
| SCOP | 1aw5 | ||||||||
| SUPERFAMILY | 1aw5 | ||||||||
| |||||||||
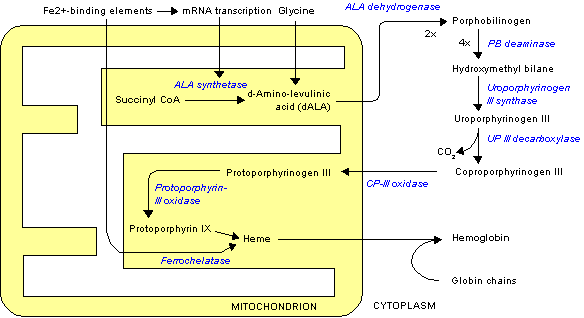
Porphobilinogen synthase (or ALA dehydratase, or aminolevulinate dehydratase) synthesizes porphobilinogen through the asymmetric condensation of two molecules of aminolevulinic acid. All natural tetrapyrroles, including hemes, chlorophylls and vitamin B12, share porphobilinogen as a common precursor.
It catalyzes the second step of the biosynthesis of porphyrin:
- 2 δ-aminolevulinic acid porphobilinogen + 2 H2O
The porphobilinogen synthase catalyzed reaction is the first common step in the biosynthesis of all biological tetrapyrroles.
Porphobilinogen synthase is the prototype morpheein.[1]
Structure
The structural basis for allosteric regulation of PBGS is modulation of a quaternary structure equilibrium between octamer and hexamer (via dimers), which is represented schematically as 6mer* ↔ 2mer* ↔ 2mer ↔ 8mer. The * represents a reorientation between two domains of each subunit that occurs in the dissociated state because it is sterically forbidden in the larger multimers.[1]
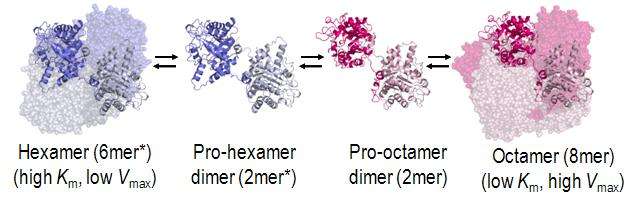
PBGS is encoded by a single gene and each PBGS multimer is composed of multiple copies of the same protein. Each PBGS subunit consists of a ~300 residue αβ-barrel domain, which houses the enzyme's active site in its center, and a >25 residue N-terminal arm domain. Allosteric regulation of PBGS can be described in terms of the orientation of the αβ-barrel domain with respect to the N-terminal arm domain.
Each N-terminal arm has up to two interactions with other subunits in a PBGS multimer. One of these interactions helps to stabilize a "closed" conformation of the active site lid. The other interaction restricts solvent access from the other end of the αβ-barrel.
In the inactive multimeric state, the N-terminal arm domain is not involved in the lid-stabilizing interaction, and in the crystal structure of the inactive assembly, the active site lid is disordered.
Allosteric regulators of PBGS
As a nearly universal enzyme with a highly conserved active site, PBGS would not be a prime target for the development of antimicrobials and/or herbicides. To the contrary, allosteric sites can be much more phylogenetically variable than active sites, thus presenting more drug development opportunities.[1]
Phylogenetic variation in PBGS allostery leads to the framing of discussion of PBGS allosteric regulation in terms of intrinsic and extrinsic factors.
Intrinsic allosteric regulators
Magnesium
The allosteric magnesium ion lies at the highly hydrated interface of two pro-octamer dimers. It appears to be easily dissociable, and it has been shown that hexamers accumulate when magnesium is removed in vitro.[2]
pH
Though it is not common to consider hydronium ions as allosteric regulators, in the case of PBGS, side chain protonation at locations other than the active site has been shown to affect the quaternary structure equilibrium, and thus to affect the rate of its catalyzed reaction as well.
Extrinsic allosteric regulators
Small molecule hexamer stabilization
Inspection of the PBGS 6mer* reveals a surface cavity that is not present in the 8mer. Small molecule binding to this phylogenetically variable cavity has been proposed to stabilize 6mer* of the targeted PBGS and consequently inhibit activity.
Such allosteric regulators are known as morphlocks because they lock PBGS in a specific morpheein form (6mer*).[3]
Deficiency
A deficiency of porphobilinogen synthase is usually acquired (rather than hereditary) and can be caused by heavy metal poisoning, especially lead poisoning, as the enzyme is very susceptible to inhibition by heavy metals.[4]
Hereditary insufficiency of porphobilinogen synthase is called porphobilinogen synthase (or ALA dehydratase) deficiency poprhyria. It is an extremely rare cause of porphyria,[5] with less than 10 cases ever reported.[6] All disease associated protein variants favor hexamer formation relative to the wild type human enzyme.[5]
 |
Lead poisoning works on the cellular level by binding to this enzyme, rendering it useless.
PBGS as the prototype morpheein
The morpheein model of allostery exemplified by PBGS adds an additional layer of understanding to potential mechanisms for regulation of protein function and complements the increased focus that the protein science community is placing on protein structure dynamics.[1]
This model illustrates how the dynamics of phenomena such as alternate protein conformations, alternate oligomeric states, and transient protein-protein interactions can be harnessed for allosteric regulation of catalytic activity.
References
- 1 2 3 4 Jaffe EK, Lawrence SH (March 2012). "Allostery and the dynamic oligomerization of porphobilinogen synthase". Arch. Biochem. Biophys. 519 (2): 144–53. PMC 3291741
 . PMID 22037356. doi:10.1016/j.abb.2011.10.010.
. PMID 22037356. doi:10.1016/j.abb.2011.10.010. - ↑ Breinig S, Kervinen J, Stith L, Wasson AS, Fairman R, Wlodawer A, Zdanov A, Jaffe EK (September 2003). "Control of tetrapyrrole biosynthesis by alternate quaternary forms of porphobilinogen synthase". Nat. Struct. Biol. 10 (9): 757–63. PMID 12897770. doi:10.1038/nsb963.
- ↑ Lawrence SH, Jaffe EK (2008). "Expanding the Concepts in Protein Structure-Function Relationships and Enzyme Kinetics: Teaching using Morpheeins". Biochem Mol Biol Educ. 36 (4): 274–283. PMC 2575429
 . PMID 19578473. doi:10.1002/bmb.20211.
. PMID 19578473. doi:10.1002/bmb.20211. - ↑ ALA dehydratase reaction, from NetBiochem at the University of Utah. Last modified 1/5/95
- 1 2 Jaffe EK, Stith L (February 2007). "ALAD porphyria is a conformational disease". Am. J. Hum. Genet. 80 (2): 329–37. PMC 1785348
 . PMID 17236137. doi:10.1086/511444.
. PMID 17236137. doi:10.1086/511444. - ↑ Overview of the Porphyrias at The Porphyrias Consortium (a part of NIH Rare Diseases Clinical Research Network (RDCRN)) Retrieved June 2011
External links
- delta-Aminolevulinic Acid Dehydratase at the US National Library of Medicine Medical Subject Headings (MeSH)
- http://www.omim.org/entry/125270?search=pbgs&highlight=pbgs
