NADH dehydrogenase (ubiquinone)
Complex I (EC 1.6.5.3) (also referred to as NADH:ubiquinone oxidoreductase or, especially in the context of the human protein, NADH dehydrogenase) is an enzyme of the respiratory chains of myriad organisms from bacteria to humans that falls under the H+ or Na+-translocating NADH Dehydrogenase (NDH) Family (TC# 3.D.1), a member of the Na+ transporting Mrp superfamily. It catalyzes the transfer of electrons from NADH to coenzyme Q10 (CoQ10) and, in eukaryotes, it is located in the inner mitochondrial membrane. NADH:ubiquinone oxidoreductases type I of bacteria and of eukaryotic mitochondria and chloroplasts couple electron transfer to the electrogenic transport of protons or Na+.[1][2] It is one of the "entry enzymes" of cellular respiration or oxidative phosphorylation in the mitochondria.[3][4]
| NADH:ubiquinone reductase (H+-translocating). | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.6.5.3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
Function
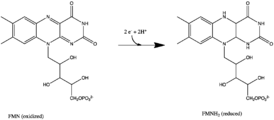

.png)
Complex I is the first enzyme of the mitochondrial electron transport chain. There are three energy-transducing enzymes in the electron transport chain - NADH:ubiquinone oxidoreductase (complex I), Coenzyme Q – cytochrome c reductase (complex III), and cytochrome c oxidase (complex IV).[5] Complex I is the largest and most complicated enzyme of the electron transport chain.[6]
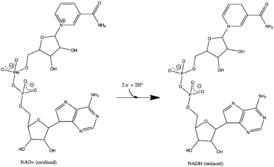
The reaction catalyzed by complex I is:
- NADH + H+ + CoQ + 4H+in→ NAD+ + CoQH2 + 4H+out
In this process, the complex translocates four protons across the inner membrane per molecule of oxidized NADH, helping to build the electrochemical potential difference used to produce ATP. Escherichia coli complex I (NADH dehydrogenase) is capable of proton translocation in the same direction to the established Δψ, showing that in the tested conditions, the coupling ion is H+.[7] Na+ transport in the opposite direction was observed, and although Na+ was not necessary for the catalytic or proton transport activities, its presence increased the latter. H+ was translocated by the Paracoccus denitrificans complex I, but in this case, H+ transport was not influenced by Na+, and Na+ transport was not observed. Possibly, the E. coli complex I has two energy coupling sites (one Na+ independent and the other Na+dependent), as observed for the Rhodothermus marinus complex I, whereas the coupling mechanism of the P. denitrificans enzyme is completely Na+ independent. It is also possible that another transporter catalyzes the uptake of Na+. Complex I energy transduction by proton pumping may not be exclusive to the R. marinus enzyme. The Na+/H+ antiport activity seems not to be a general property of complex I.[7] However, the existence of Na+-translocating activity of the complex I is still in question.
The reaction can be reversed – referred to as aerobic succinate-supported NAD+ reduction by ubiquinol – in the presence of a high membrane potential, but the exact catalytic mechanism remains unknown. Driving force of this reaction is a potential across the membrane which can be maintained eitehr by ATP-hydrolysis or by complexes III and IV during succinate oxidation.[8]
Complex I may have a role in triggering apoptosis.[9] In fact, there has been shown to be a correlation between mitochondrial activities and programmed cell death (PCD) during somatic embryo development.[10]
Mechanism
Overall mechanism
All redox reactions take place in the hydrophilic domain of complex I. NADH initially binds to complex I, and transfers two electrons to the flavin mononucleotide (FMN) prosthetic group of the enzyme, creating FMNH2. The electron acceptor – the isoalloxazine ring – of FMN is identical to that of FAD. The electrons are then transferred through the FMN via a series of iron-sulfur (Fe-S) clusters[how many?], and finally to coenzyme Q10 (ubiquinone). This electron flow changes the redox state of the protein, inducing conformational changes of the protein which alters the pK values of ionizable side chain, and causes four hydrogen ions to be pumped out of the mitochondrial matrix.[11] Ubiquinone (CoQ) accepts two electrons to be reduced to ubiquinol (CoQH2).[5]
Electron transfer mechanism
The proposed pathway for electron transport prior to ubiquinone reduction is as follows: NADH – FMN – N3 – N1b – N4 – N5 – N6a – N6b – N2 – Q, where Nx is a labelling convention for iron sulfur clusters.[12] The high reduction potential of the N2 cluster and the relative proximity of the other clusters in the chain enable efficient electron transfer over long distance in the protein (with transfer rates from NADH to N2 iron-sulfur cluster of about 100 μs).[13][14]
The equilibrium dynamics of Complex I are primarily driven by the quinone redox cycle. In conditions of high proton motive force (and accordingly, a ubiquinol-concentrated pool), the enzyme runs in the reverse direction. Ubiquinol is oxidized to ubiquinone, and the resulting released protons reduce the proton motive force.[15]
Proton translocation mechanism
The coupling of proton translocation and electron transport in Complex I is currently proposed as being indirect (long range conformational changes) as opposed to direct (redox intermediates in the hydrogen pumps as in heme groups of Complexes III and IV).[12] The architecture of the hydrophobic region of complex I shows multiple proton transporters that are mechanically interlinked. The three central components believed to contribute to this long-range conformational change event are the pH-coupled N2 iron-sulfur cluster, the quinone reduction, and the transmembrane helix subunits of the membrane arm. Transduction of conformational changes to drive the transmembrane transporters linked by a 'connecting rod' during the reduction of ubiquinone can account for two or three of the four protons pumped per NADH oxidized. The remaining proton must be pumped by direct coupling at the ubiquinone-binding site. It is proposed that direct and indirect coupling mechanisms account for the pumping of the four protons.[16]
The N2 cluster's proximity to a nearby cysteine residue results in a conformational change upon reduction in the nearby helicases, leading to small but important changes in the overall protein conformation.[17] Further electron paramagnetic resonance studies of the electron transfer have demonstrated that most of the energy that is released during the subsequent CoQ reduction is on the final ubiquinol formation step from semiquinone, providing evidence for the "single stroke" H+ translocation mechanism (i.e. all four protons move across the membrane at the same time).[15][18] Alternative theories suggest a "two stroke mechanism" where each reduction step (semiquinone and ubiquinol) results in a stroke of two protons entering the intermembrane space.[19][20]
The resulting ubiquinol localized to the membrane domain interacts with negatively charged residues in the membrane arm, stabilizing conformational changes.[12] An antiporter mechanism (Na+/H+ swap) has been proposed using evidence of conserved Asp residues in the membrane arm.[21] The presence of Lys, Glu, and His residues enable for proton gating (a protonation followed by deprotonation event across the membrane) driven by the pKa of the residues.[12]
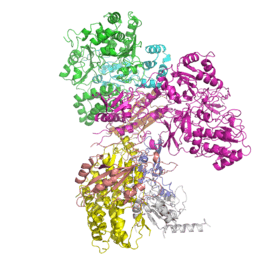
Composition and structure
NADH:ubiquinone oxidoreductase is the largest of the respiratory complexes. In mammals, the enzyme contains 44 separate water soluble peripheral membrane proteins, which are anchored to the integral membrane constituents. Of particular functional importance are the flavin prosthetic group (FMN) and eight iron-sulfur clusters (FeS). Of the 44 subunits, seven are encoded by the mitochondrial genome.[22][23][24]
The structure is an "L" shape with a long membrane domain (with around 60 trans-membrane helices) and a hydrophilic (or peripheral) domain, which includes all the known redox centres and the NADH binding site. The structure of the eukaryotic complex is not well characterised. However, the Sazanov group succeeded in solving the structures of the complex I hydrophilic domain from the bacterium Thermus thermophilus (PDB: 2FUG) [25] and complex I membrane domains from both the E. coli (PDB: 3rko) and T. thermophilus (PDB: 4HE8) enzymes. In February 2013 the structure of an entire, intact complex I (from T. thermophilus) was published for the first time, again by the Sazanov group (PDB: 4HEA). All thirteen of the E. coli proteins, which comprise NADH dehydrogenase I, are encoded within the nuo operon, and are homologous to mitochondrial complex I subunits. The antiporter-like subunits NuoL/M/N each contains 14 conserved transmembrane (TM) helices. Two of them are discontinuous, but subunit NuoL contains a 110 Å long amphipathic α-helix, spanning the entire length of the domain. The subunit, NuoL, is related to Na+/ H+ antiporters of TC# 2.A.63.1.1 (PhaA and PhaD).
Three of the conserved, membrane-bound subunits in NADH dehydrogenase are related to each other, and to Mrp sodium-proton antiporters. Structural analysis of two prokaryotic complexes I revealed that the three subunits each contain fourteen transmembrane helices that overlay in structural alignments: the translocation of three protons may be coordinated by a lateral helix connecting them.[26]
Complex I contains a ubiquinone binding pocket at the interface of the 49-kDa and PSST subunits. Close to iron-sulfur cluster N2, the proposed immediate electron donor for ubiquinone, a highly conserved tyrosine constitutes a critical element of the quinone reduction site. A possible quinone exchange path leads from cluster N2 to the N-terminal beta-sheet of the 49-kDa subunit.[27] All 45 subunits of the bovine NDHI have been sequenced.[28][29] Each complex contains noncovalently bound FMN, coenzyme Q and several iron-sulfur centers. The bacterial NDHs have 8-9 iron-sulfur centers.
A recent study by Roessler et al. (2010) used electron paramagnetic resonance (EPR) spectra and double electron-electron resonance (DEER) to determine the path of electron transfer through the iron-sulfur complexes, which are located in the hydrophilic domain. Seven of these clusters form a chain from the flavin to the quinone binding sites; the eighth cluster is located on the other side of the flavin, and its function is unknown. The EPR and DEER results suggest an alternating or “roller-coaster” potential energy profile for the electron transfer between the active sites and along the iron-sulfur clusters, which can optimize the rate of electron travel and allow efficient energy conversion in complex I.[30]
A simulational study by Hayashi and Stuchebrukhov further identified the electron tunneling pathways in atomic resolution based on the tunneling current theory. The distinct pathways between neighboring Fe/S clusters primarily consist of two cysteine ligands and one additional key residue, which was supported by sensitivity of simulated electron transfer rates to their mutations and their conservation among various complex I homologues from simple bacteria to human beings. This result shows that the crucial part of complex I developed for optimal efficiency with specific key residues during early stages of the biological evolution and has been conserved since then. Internal water between protein subunits was identified as an essential mediator enhancing the overall electron transfer rate to achieve physiologically significant value.[31][32]
| # | Human/Bovine subunit | Human protein | Protein description (UniProt) | Pfam family with Human protein | |
|---|---|---|---|---|---|
| Core Subunitsa | |||||
| 1 | NDUFS7 / PSST / NUKM | NDUS7_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 7, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF01058 | |
| 2 | NDUFS8 / TYKY / NUIM | NDUS8_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 8, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF12838 | |
| 3 | NDUFV2 / 24kD / NUHMc | NDUV2_HUMAN | NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF01257 | |
| 4 | NDUFS3 / 30kD / NUGM | NDUS3_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 3, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF00329 | |
| 5 | NDUFS2 / 49kD / NUCM | NDUS2_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 2, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF00346 | |
| 6 | NDUFV1 / 51kD / NUBM | NDUV1_HUMAN | NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF01512 | |
| 7 | NDUFS1 / 75kD / NUAM | NDUS1_HUMAN | NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial EC 1.6.5.3 EC 1.6.99.3 | Pfam PF00384 | |
| 8 | ND1 / NU1M | NU1M_HUMAN | NADH-ubiquinone oxidoreductase chain 1 EC 1.6.5.3 | Pfam PF00146 | |
| 9 | ND2 / NU2M | NU2M_HUMAN | NADH-ubiquinone oxidoreductase chain 2 EC 1.6.5.3 | Pfam PF00361, Pfam PF06444 | |
| 10 | ND3 / NU3M | NU3M_HUMAN | NADH-ubiquinone oxidoreductase chain 3 EC 1.6.5.3 | Pfam PF00507 | |
| 11 | ND4 / NU4M | NU4M_HUMAN | NADH-ubiquinone oxidoreductase chain 4 EC 1.6.5.3 | Pfam PF01059,Pfam PF00361 | |
| 12 | ND4L / NULM | NU4LM_HUMAN | NADH-ubiquinone oxidoreductase chain 4L EC 1.6.5.3 | Pfam PF00420 | |
| 13 | ND5 / NU5M | NU5M_HUMAN | NADH-ubiquinone oxidoreductase chain 5 EC 1.6.5.3 | Pfam PF00361, Pfam PF06455, Pfam PF00662 | |
| 14 | ND6 / NU6M | NU6M_HUMAN | NADH-ubiquinone oxidoreductase chain 6 EC 1.6.5.3 | Pfam PF00499 | |
| Core accessory subunitsb | |||||
| 15 | NDUFS6 / 13A | NDUS6_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 6, mitochondrial | Pfam PF10276 | |
| 16 | NDUFA12 / B17.2 | NDUAC_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 12 | Pfam PF05071 | |
| 17 | NDUFS4 / AQDQ | NDUS4_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 4, mitochondrial | Pfam PF04800 | |
| 18 | NDUFA9 / 39kDa | NDUA9_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | Pfam PF01370 | |
| 19 | NDUFAB1 / ACPM | ACPM_HUMAN | Acyl carrier protein, mitochondrial | Pfam PF00550 | |
| 20 | NDUFA2 / B8 | NDUA2_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 | Pfam PF05047 | |
| 21 | NDUFA1 / MFWE | NDUA1_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 1 | Pfam PF15879 | |
| 22 | NDUFB3 / B12 | NDUB3_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 3 | Pfam PF08122 | |
| 23 | NDUFA5 / AB13 | NDUA5_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | Pfam PF04716 | |
| 24 | NDUFA6 / B14 | NDUA6_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 6 | Pfam PF05347 | |
| 25 | NDUFA11 / B14.7 | NDUAB_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 11 | Pfam PF02466 | |
| 26 | NDUFB11 / ESSS | NDUBB_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 11, mitochondrial | Pfam PF10183 | |
| 27 | NDUFS5 / PFFD | NDUS5_HUMAN | NADH dehydrogenase [ubiquinone] iron-sulfur protein 5 | Pfam PF10200 | |
| 28 | NDUFB4 / B15 | NDUB4_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 4 | Pfam PF07225 | |
| 29 | NDUFA13 /A13 | NDUAD_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13 | Pfam PF06212 | |
| 30 | NDUFB7 / B18 | NDUB7_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 | Pfam PF05676 | |
| 31 | NDUFA8 / PGIV | NDUA8_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 8 | Pfam PF06747 | |
| 32 | NDUFB9 / B22 | NDUB9_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 | Pfam PF05347 | |
| 33 | NDUFB10 / PDSW | NDUBA_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10 | Pfam PF10249 | |
| 34 | NDUFB8 / ASHI | NDUB8_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial | Pfam PF05821 | |
| 35 | NDUFC2 / B14.5B | NDUC2_HUMAN | NADH dehydrogenase [ubiquinone] 1 subunit C2 | Pfam PF06374 | |
| 36 | NDUFB2 / AGGG | NDUB2_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 2, mitochondrial | Pfam PF14813 | |
| 37 | NDUFA7 / B14.5A | NDUA7_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7 | Pfam PF07347 | |
| 38 | NDUFA3 / B9 | NDUA3_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 3 | Pfam PF14987 | |
| 39 | NDUFA4 / MLRQc | NDUA4_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4 | Pfam PF06522 | |
| 40 | NDUFB5 / SGDH | NDUB5_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 5, mitochondrial | Pfam PF09781 | |
| 41 | NDUFB1 / MNLL | NDUB1_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 1 | Pfam PF08040 | |
| 42 | NDUFC1 / KFYI | NDUC1_HUMAN | NADH dehydrogenase [ubiquinone] 1 subunit C1, mitochondrial | Pfam PF15088 | |
| 43 | NDUFA10 / 42kD | NDUAA_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial | Pfam PF01712 | |
| 44 | NDUFA4L2 | NUA4L_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 4-like 2 | Pfam PF15880 | |
| 45 | NDUFV3 | NDUV3_HUMAN | NADH dehydrogenase [ubiquinone] flavoprotein 3, 10kDa | - | |
| 46 | NDUFB6 | NDUB6_HUMAN | NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 6 | Pfam PF09782 | |
| Assembly factor proteins[34] | |||||
| 47 | NDUFAF1c | CIA30_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, assembly factor 1 | Pfam PF08547 | |
| 48 | NDUFAF2 | MIMIT_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, assembly factor 2 | Pfam PF05071 | |
| 49 | NDUFAF3 | NDUF3_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3 | Pfam PF05071 | |
| 50 | NDUFAF4 | NDUF4_HUMAN | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex, assembly factor 4 | Pfam PF06784 | |
Notes:
- a Found in all species except fungi
- b May or may not be present in any species
- c Found in fungal species such as Schizosaccharomyces pombe
Inhibitors
The best-known inhibitor of complex I is rotenone (commonly used as an organic pesticide). Rotenone and rotenoids are isoflavonoids occurring in several genera of tropical plants such as Antonia (Loganiaceae), Derris and Lonchocarpus (Faboideae, Fabaceae). There have been reports of the indigenous people of French Guiana using rotenone-containing plants to fish - due to its ichthyotoxic effect - as early as the 17th century.[35] Rotenone binds to the ubiquinone binding site of complex I as well as piericidin A, another potent inhibitor with a close structural homologue to ubiquinone.
Acetogenins from Annonaceae are even more potent inhibitors of complex I. They cross-link to the ND2 subunit, which suggests that ND2 is essential for quinone-binding.[3] Interestingly, Rolliniastatin-2, an acetogenin, is the first complex I inhibitor found that does not share the same binding site as rotenone.[36]
Despite more than 50 years of study of complex I, no inhibitors blocking the electron flow inside the enzyme have been found. Hydrophobic inhibitors like rotenone or piericidin most likely disrupt the electron transfer between the terminal FeS cluster N2 and ubiquinone. It has been shown that long-term systemic inhibition of complex I by rotenone can induce selective degeneration of dopaminergic neurons.[37]
Complex I is also blocked by adenosine diphosphate ribose – a reversible competitive inhibitor of NADH oxidation – by binding to the enzyme at the nucleotide binding site.[38] Both hydrophilic NADH and hydrophobic ubiquinone analogs act at the beginning and the end of the internal electron-transport pathway, respectively.
The antidiabetic drug Metformin has been shown to induce a mild and transient inhibition of the mitochondrial respiratory chain complex I, and this inhibition appears to play a key role in its mechanism of action.[39]
Inhibition of complex I has been implicated in hepatotoxicity associated with a variety of drugs, for instance flutamide and nefazodone.[40]
Active/deactive transition
The catalytic properties of eukaryotic complex I are not simple. Two catalytically and structurally distinct forms exist in any given preparation of the enzyme: one is the fully competent, so-called “active” A-form and the other is the catalytically silent, dormant, “deactive”, D-form. After exposure of idle enzyme to elevated, but physiological temperatures (>30 °C) in the absence of substrate, the enzyme converts to the D-form. This form is catalytically incompetent but can be activated by the slow reaction (k~4 min−1) of NADH oxidation with subsequent ubiquinone reduction. After one or several turnovers the enzyme becomes active and can catalyse physiological NADH:ubiquinone reaction at a much higher rate (k~104 min−1). In the presence of divalent cations (Mg2+, Ca2+), or at alkaline pH the activation takes much longer.
The high activation energy (270 kJ/mol) of the deactivation process indicates the occurrence of major conformational changes in the organisation of the complex I. However, until now, the only conformational difference observed between these two forms is the number of cysteine residues exposed at the surface of the enzyme. Treatment of the D-form of complex I with the sulfhydryl reagents N-Ethylmaleimide or DTNB irreversibly blocks critical cysteine residue(s), abolishing the ability of the enzyme to respond to activation, thus inactivating it irreversibly. The A-form of complex I is insensitive to sulfhydryl reagents.
It was found that these conformational changes may have a very important physiological significance. The deactive, but not the active form of complex I was susceptible to inhibition by nitrosothiols and peroxynitrite.[41] It is likely that transition from the active to the inactive form of complex I takes place during pathological conditions when the turnover of the enzyme is limited at physiological temperatures, such as during hypoxia, or when the tissue nitric oxide:oxygen ratio increases (i.e. metabolic hypoxia).[42]
Production of superoxide
Recent investigations suggest that complex I is a potent source of reactive oxygen species.[43] Complex I can produce superoxide (as well as hydrogen peroxide), through at least two different pathways. During forward electron transfer, only very small amounts of superoxide are produced (probably less than 0.1% of the overall electron flow).[43][44]
During reverse electron transfer, complex I might be the most important site of superoxide production within mitochondria, with up to 5% of electrons being diverted to superoxide formation. Reverse electron transfer, the process by which electrons from the reduced ubiquinol pool (supplied by succinate dehydrogenase, glycerol-3-phosphate dehydrogenase, or dihydro-oorotate dehydrogenase in mammalian mitochondria) pass through complex I to reduce NAD+ to NADH, driven by the inner mitochondrial membrane potential electric potential. Although it is not precisely known under what pathological conditions reverse-electron transfer would occur in vivo, in vitro experiments indicate that it can be a very potent source of superoxide when succinate concentrations are high and oxaloacetate or malate concentrations are low.[45]
Superoxide is a reactive oxygen species that contributes to cellular oxidative stress and is linked to neuromuscular diseases and aging.[46] NADH dehdyrogenase produces superoxide by transferring one electron from FMNH2 to oxygen (O2). The radical flavin leftover is unstable, and transfers the remaining electron to the iron-sulfur centers. Interestingly, it is the ratio of NADH to NAD+ that determines the rate of superoxide formation.[47]
Pathology
Mutations in the subunits of complex I can cause mitochondrial diseases, including Leigh syndrome. Point mutations in various complex I subunits derived from mitochondrial DNA (mtDNA) can also result in Leber's Hereditary Optic Neuropathy. There is some evidence that complex I defects may play a role in the etiology of Parkinson's disease, perhaps because of reactive oxygen species (complex I can, like complex III, leak electrons to oxygen, forming highly toxic superoxide).
Although the exact etiology of Parkinson’s disease is unclear, it is likely that mitochondrial dysfunction, along with proteasome inhibition and environmental toxins, may play a large role. In fact, the inhibition of complex I has been shown to cause the production of peroxides and a decrease in proteasome activity, which may lead to Parkinson’s disease.[48] Additionally, Esteves et al. (2010) found that cell lines with Parkinson’s disease show increased proton leakage in complex I, which causes decreased maximum respiratory capacity.[49]
Recent studies have examined other roles of complex I activity in the brain. Andreazza et al. (2010) found that the level of complex I activity was significantly decreased in patients with bipolar disorder, but not in patients with depression or schizophrenia. They found that patients with bipolar disorder showed increased protein oxidation and nitration in their prefrontal cortex. These results suggest that future studies should target complex I for potential therapeutic studies for bipolar disorder.[50] Similarly, Moran et al. (2010) found that patients with severe complex I deficiency showed decreased oxygen consumption rates and slower growth rates. However, they found that mutations in different genes in complex I lead to different phenotypes, thereby explaining the variations of pathophysiological manifestations of complex I deficiency.[51]
Exposure to pesticides can also inhibit complex I and cause disease symptoms. For example, chronic exposure to low levels of dichlorvos, an organophosphate used as a pesticide, has been shown to cause liver dysfunction. This occurs because dichlorvos alters complex I and II activity levels, which leads to decreased mitochondrial electron transfer activities and decreased ATP synthesis.[52]
Genes
The following is a list of humans genes that encode components of complex I:
- NADH dehydrogenase (ubiquinone) 1 alpha subcomplex
- NDUFA1 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa
- NDUFA2 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa
- NDUFA3 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa
- NDUFA4 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa
- NDUFA4L – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like
- NDUFA4L2 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2
- NDUFA5 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa
- NDUFA6 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa
- NDUFA7 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa
- NDUFA8 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa
- NDUFA9 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa
- NDUFA10 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa
- NDUFA11 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa
- NDUFA12 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12
- NDUFA13 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13
- NDUFAB1 – NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa
- NDUFAF1 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1
- NDUFAF2 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2
- NDUFAF3 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3
- NDUFAF4 – NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 4
- NADH dehydrogenase (ubiquinone) 1 beta subcomplex
- NDUFB1 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 1, 7kDa
- NDUFB2 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa
- NDUFB3 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa
- NDUFB4 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa
- NDUFB5 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa
- NDUFB6 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6, 17kDa
- NDUFB7 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7, 18kDa
- NDUFB8 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa
- NDUFB9 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa
- NDUFB10 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa
- NDUFB11 – NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 11, 17.3kDa
- NADH dehydrogenase (ubiquinone) 1, subcomplex unknown
- NADH dehydrogenase (ubiquinone) Fe-S protein
- NDUFS1 – NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase)
- NDUFS2 – NADH dehydrogenase (ubiquinone) Fe-S protein 2, 49kDa (NADH-coenzyme Q reductase)
- NDUFS3 – NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase)
- NDUFS4 – NADH dehydrogenase (ubiquinone) Fe-S protein 4, 18kDa (NADH-coenzyme Q reductase)
- NDUFS5 – NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase)
- NDUFS6 – NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase)
- NDUFS7 – NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase)
- NDUFS8 – NADH dehydrogenase (ubiquinone) Fe-S protein 8, 23kDa (NADH-coenzyme Q reductase)
- NADH dehydrogenase (ubiquinone) flavoprotein 1
- mitochondrially encoded NADH dehydrogenase subunit
- MT-ND1 - mitochondrially encoded NADH dehydrogenase subunit 1
- MT-ND2 - mitochondrially encoded NADH dehydrogenase subunit 2
- MT-ND3 - mitochondrially encoded NADH dehydrogenase subunit 3
- MT-ND4 - mitochondrially encoded NADH dehydrogenase subunit 4
- MT-ND4L - mitochondrially encoded NADH dehydrogenase subunit 4L
- MT-ND5 - mitochondrially encoded NADH dehydrogenase subunit 5
- MT-ND6 - mitochondrially encoded NADH dehydrogenase subunit 6
References
- ↑ Brandt, Ulrich (2006-01-01). "Energy converting NADH:quinone oxidoreductase (complex I)". Annual Review of Biochemistry. 75: 69–92. ISSN 0066-4154. PMID 16756485. doi:10.1146/annurev.biochem.75.103004.142539.
- ↑ Gemperli, Anja C.; Schaffitzel, Christiane; Jakob, Claude; Steuber, Julia (2007-11-01). "Transport of Na(+) and K (+) by an antiporter-related subunit from the Escherichia coli NADH dehydrogenase I produced in Saccharomyces cerevisiae". Archives of Microbiology. 188 (5): 509–521. ISSN 0302-8933. PMID 17583799. doi:10.1007/s00203-007-0272-3.
- 1 2 Nakamaru-Ogiso E, Han H, Matsuno-Yagi A, Keinan E, Sinha SC, Yagi T, Ohnishi T (January 2010). "The ND2 subunit is labeled by a photoaffinity analogue of asimicin, a potent complex I inhibitor.". FEBS Letters. 584 (5): 883–8. PMC 2836797
 . PMID 20074573. doi:10.1016/j.febslet.2010.01.004.
. PMID 20074573. doi:10.1016/j.febslet.2010.01.004. - ↑ "Protein Data Bank in Europe: Bringing Structure to Biology". Retrieved 30 March 2014.
- 1 2 Berg, J; Tymoczko, J; L Stryer (2006). Biochemistry (6th ed.). New York: WH Freeman & Company. pp. 509–513.
- ↑ Brandt, U (2006). "Energy converting NADH:quinone oxidoreductase (complex I)". Annual Review of Biochemistry. 75: 69–92. PMID 16756485. doi:10.1146/annurev.biochem.75.103004.142539.
- 1 2 Batista, Ana P.; Pereira, Manuela M. (2011-03-01). "Sodium influence on energy transduction by complexes I from Escherichia coli and Paracoccus denitrificans". Biochimica et Biophysica Acta. 1807 (3): 286–292. ISSN 0006-3002. PMID 21172303. doi:10.1016/j.bbabio.2010.12.008.
- ↑ Grivennikova VG, Kotlyar AB, Karliner JS, Cecchini G, Vinogradov AD (August 2007). "Redox-dependent change of nucleotide affinity to the active site of the mammalian complex I.". Biochemistry. 46 (38): 10971–8. PMC 2258335
 . PMID 17760425. doi:10.1021/bi7009822.
. PMID 17760425. doi:10.1021/bi7009822. - ↑ Chomova M, Racay P (March 2010). "Mitochondrial complex I in the network of known and unknown facts.". General Physiology and Biophysics. 29 (1): 3–11. PMID 20371875. doi:10.4149/gpb_2010_01_3.
- ↑ Petrussa E, Bertolini A, Casolo V, Krajnáková J, Macrì F, Vianello A (October 2009). "Mitochondrial bioenergetics linked to the manifestation of programmed cell death during somatic embryogenesis of Abies alba.". Planta. 231 (1): 93–107. PMID 19834734. doi:10.1007/s00425-009-1028-x.
- ↑ Donald J. Voet; Judith G. Voet; Charlotte W. Pratt (2008). "Chapter 18, Mitochondrial ATP synthesis". Principles of Biochemistry, 3rd Edition. Wiley. p. 608. ISBN 978-0-470-23396-2.
- 1 2 3 4 Sazanov, Leonid A. (2015-06-01). "A giant molecular proton pump: structure and mechanism of respiratory complex I". Nature Reviews Molecular Cell Biology. 16 (6): 375–388. ISSN 1471-0072. PMID 25991374. doi:10.1038/nrm3997.
- ↑ Ohnishi, T (1998). "Iron–sulfur clusters/semiquinones in complex I". Biochim. Biophys. Acta. 1364: 186–206. doi:10.1016/s0005-2728(98)00027-9.
- ↑ Bridges, H. R.; Bill, E.; Hirst, J. (2012). "Mossbauer spectroscopy on respiratory complex I: the iron–sulfur cluster ensemble in the NADH-reduced enzyme is partially oxidized". Biochemistry. 51: 149–158. doi:10.1021/bi201644x.
- 1 2 Efremov, Rouslan G.; Sazanov, Leonid A. (2012-10-01). "The coupling mechanism of respiratory complex I - a structural and evolutionary perspective". Biochimica et Biophysica Acta. 1817 (10): 1785–1795. ISSN 0006-3002. PMID 22386882. doi:10.1016/j.bbabio.2012.02.015.
- ↑ Treberg, Jason R.; Quinlan, Casey L.; Brand, Martin D. (2011-08-05). "Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I)". The Journal of Biological Chemistry. 286 (31): 27103–27110. ISSN 1083-351X. PMC 3149303
 . PMID 21659507. doi:10.1074/jbc.M111.252502.
. PMID 21659507. doi:10.1074/jbc.M111.252502. - ↑ Berrisford, J. M.; Sazanov, L. A. (2009). "Structural basis for the mechanism of respiratory complex I". J. Biol. Chem. 284: 29773–29783. PMC 2785608
 . PMID 19635800. doi:10.1074/jbc.m109.032144.
. PMID 19635800. doi:10.1074/jbc.m109.032144. - ↑ Baranova, Ekaterina A.; Morgan, David J.; Sazanov, Leonid A. (2007-08-01). "Single particle analysis confirms distal location of subunits NuoL and NuoM in Escherichia coli complex I". Journal of Structural Biology. 159 (2): 238–242. ISSN 1047-8477. PMID 17360196. doi:10.1016/j.jsb.2007.01.009.
- ↑ Brandt, Ulrich (2011-10-01). "A two-state stabilization-change mechanism for proton-pumping complex I". Biochimica et Biophysica Acta. 1807 (10): 1364–1369. ISSN 0006-3002. PMID 21565159. doi:10.1016/j.bbabio.2011.04.006.
- ↑ Zickermann, Volker; Wirth, Christophe; Nasiri, Hamid; Siegmund, Karin; Schwalbe, Harald; Hunte, Carola; Brandt, Ulrich (2015-01-02). "Structural biology. Mechanistic insight from the crystal structure of mitochondrial complex I". Science. 347 (6217): 44–49. ISSN 1095-9203. PMID 25554780. doi:10.1126/science.1259859.
- ↑ Hunte, Carola; Screpanti, Emanuela; Venturi, Miro; Rimon, Abraham; Padan, Etana; Michel, Hartmut (2005-06-30). "Structure of a Na+/H+ antiporter and insights into mechanism of action and regulation by pH". Nature. 435 (7046): 1197–1202. ISSN 1476-4687. PMID 15988517. doi:10.1038/nature03692.
- ↑ Voet, Judith G.; Voet, Donald (2004). Biochemistry (3rd ed.). New York: J. Wiley & Sons. pp. 813–826. ISBN 0-471-19350-X.
- ↑ Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J, Walker JE (October 2006). "Bovine complex I is a complex of 45 different subunits". J. Biol. Chem. 281 (43): 32724–7. PMID 16950771. doi:10.1074/jbc.M607135200.
- ↑ Balsa E, Marco R, Perales-Clemente E, Szklarczyk R, Calvo E, Landázuri MO, Enríquez JA (September 2012). "NDUFA4 is a subunit of complex IV of the mammalian electron transport chain". Cell Metab. 16 (3): 378–86. PMID 22902835. doi:10.1016/j.cmet.2012.07.015.
- ↑ Sazanov LA, Hinchliffe P (March 2006). "Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus". Science. 311 (5766): 1430–6. PMID 16469879. doi:10.1126/science.1123809.
- ↑ Efremov, Rouslan G.; Baradaran, Rozbeh; Sazanov, Leonid A. (2010-05-27). "The architecture of respiratory complex I". Nature. 465 (7297): 441–445. ISSN 1476-4687. PMID 20505720. doi:10.1038/nature09066.
- ↑ Tocilescu, Maja A.; Zickermann, Volker; Zwicker, Klaus; Brandt, Ulrich (2010-12-01). "Quinone binding and reduction by respiratory complex I". Biochimica et Biophysica Acta. 1797 (12): 1883–1890. ISSN 0006-3002. PMID 20493164. doi:10.1016/j.bbabio.2010.05.009.
- ↑ Cardol, Pierre; Vanrobaeys, Frank; Devreese, Bart; Van Beeumen, Jozef; Matagne, René F.; Remacle, Claire (2004-10-04). "Higher plant-like subunit composition of mitochondrial complex I from Chlamydomonas reinhardtii: 31 conserved components among eukaryotes". Biochimica et Biophysica Acta. 1658 (3): 212–224. ISSN 0006-3002. PMID 15450959. doi:10.1016/j.bbabio.2004.06.001.
- ↑ Gabaldón, Toni; Rainey, Daphne; Huynen, Martijn A. (2005-05-13). "Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I)". Journal of Molecular Biology. 348 (4): 857–870. ISSN 0022-2836. PMID 15843018. doi:10.1016/j.jmb.2005.02.067.
- ↑ Roessler MM, King MS, Robinson AJ, Armstrong FA, Harmer J, Hirst J (February 2010). "Direct assignment of EPR spectra to structurally defined iron-sulfur clusters in complex I by double electron-electron resonance..". Proceedings of the National Academy of Sciences of the United States of America. 107 (5): 1930–5. PMC 2808219
 . PMID 20133838. doi:10.1073/pnas.0908050107.
. PMID 20133838. doi:10.1073/pnas.0908050107. - ↑ Hayashi T.; Stuchebrukhov A. (November 2010). "Electron Tunneling in Respiratory Complex I..". Proceedings of the National Academy of Sciences of the United States of America. 107 (45): 19157–19162. PMC 2984193
 . PMID 20974925. doi:10.1073/pnas.1009181107.
. PMID 20974925. doi:10.1073/pnas.1009181107. - ↑ Electron Tunneling Pathways in Respiratory Complex I: 3D Rotating Movie on YouTube
- ↑ Cardol P (2011). "Mitochondrial NADH:ubiquinone oxidoreductase (complex I) in eukaryotes: a highly conserved subunit composition highlighted by mining of protein databases.". Biochim Biophys Acta. 1807 (11): 1390–7. PMID 21749854. doi:10.1016/j.bbabio.2011.06.015.
- ↑ Ogilvie I, Kennaway NG, Shoubridge EA (2005). "A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy.". J Clin Invest. 115 (10): 2784–92. PMC 1236688
 . PMID 16200211. doi:10.1172/JCI26020.
. PMID 16200211. doi:10.1172/JCI26020. - ↑ Moretti C, Grenand P (September 1988). "[The "nivrées", or ichthyotoxic plants of French Guyana]". J Ethnopharmacol (in French). 6 (2): 139–60. PMID 7132401. doi:10.1016/0378-8741(82)90002-2.
- ↑ Degli Esposti M, Ghelli A, Ratta M, Cortes D, Estornell E (July 1994). "Natural substances (acetogenins) from the family Annonaceae are powerful inhibitors of mitochondrial NADH dehydrogenase (complex I)". The Biochemical Journal. 301: 161–7. PMC 1137156
 . PMID 8037664.
. PMID 8037664. - ↑ Watabe M, Nakaki T (July 2008). "Mitochondrial complex I inhibitor rotenone inhibits and redistributes vesicular monoamine transporter 2 via nitration in human dopaminergic SH-SY5Y cells". Molecular Pharmocology. 74 (4): 933–40. PMID 18599602. doi:10.1124/mol.108.048546.
- ↑ Zharova TV, Vinogradov AD (July 1997). "A competitive inhibition of the mitochondrial NADH-ubiquinone oxidoreductase (complex I) by ADP-ribose". Biochimica et Biophysica Acta. 1320 (3): 256–64. PMID 9230920. doi:10.1016/S0005-2728(97)00029-7.
- ↑ Viollet B, Guigas B, Sanz Garcia N, Leclerc J, Foretz M, Andreelli F (March 2012). "Cellular and molecular mechanisms of metformin: an overview". Clinical Science (London). 122 (6): 253–70. PMC 3398862
 . PMID 22117616. doi:10.1042/CS20110386.
. PMID 22117616. doi:10.1042/CS20110386. - ↑ Nadanaciva, Sashi; Will, Yvonne (2011). "New Insights in Drug-Induced Mitochondrial Toxicity". Current Pharmaceutical Design. 17 (20): 2100–2112. ISSN 1381-6128. doi:10.2174/138161211796904795.
- ↑ Galkin A, Moncada S (December 2007). "S-nitrosation of mitochondrial complex I depends on its structural conformation". J. Biol. Chem. 282 (52): 37448–53. PMID 17956863. doi:10.1074/jbc.M707543200.
- ↑ Moncada S, Erusalimsky JD (March 2002). "Does nitric oxide modulate mitochondrial energy generation and apoptosis?". Nat. Rev. Mol. Cell Biol. 3 (3): 214–20. PMID 11994742. doi:10.1038/nrm762.
- 1 2 Murphy MP (January 2009). "How mitochondria produce reactive oxygen species". Biochem. J. 417 (1): 1–13. PMC 2605959
 . PMID 19061483. doi:10.1042/BJ20081386.
. PMID 19061483. doi:10.1042/BJ20081386. - ↑ Hansford RG, Hogue BA, Mildaziene V (February 1997). "Dependence of H2O2 formation by rat heart mitochondria on substrate availability and donor age". J. Bioenerg. Biomembr. 29 (1): 89–95. PMID 9067806. doi:10.1023/A:1022420007908.
- ↑ Muller FL, Liu Y, Abdul-Ghani MA, Lustgarten MS, Bhattacharya A, Jang YC, Van Remmen H (January 2008). "High rates of superoxide production in skeletal-muscle mitochondria respiring on both complex I- and complex II-linked substrates". Biochem. J. 409 (2): 491–9. PMID 17916065. doi:10.1042/BJ20071162.
- ↑ Esterházy D, King MS, Yakovlev G, Hirst J (March 2008). "Production of reactive oxygen species by complex I (NADH:ubiquinone oxidoreductase) from Escherichia coli and comparison to the enzyme from mitochondria". Biochemistry. 47 (12): 3964–71. PMID 18307315. doi:10.1021/bi702243b.
- ↑ Kussmaul L, Hirst J (May 2006). "The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria". Proceedings of the National Academy of Sciences of the United States of America. 103 (20): 7607–12. PMC 1472492
 . PMID 16682634. doi:10.1073/pnas.0510977103.
. PMID 16682634. doi:10.1073/pnas.0510977103. - ↑ Chou AP, Li S, Fitzmaurice AG, Bronstein JM (April 2010). "Mechanisms of rotenone-induced proteasome inhibition". Neurotoxicology. 31 (4): 367–72. PMC 2885979
 . PMID 20417232. doi:10.1016/j.neuro.2010.04.006.
. PMID 20417232. doi:10.1016/j.neuro.2010.04.006. - ↑ Esteves AR, Lu J, Rodova M, Onyango I, Lezi E, Dubinsky R, Lyons KE, Pahwa R, Burns JM, Cardoso SM, Swerdlow RH (February 2010). "Mitochondrial respiration and respiration-associated proteins in cell lines created through Parkinson's subject mitochondrial transfer". Journal of Neurochemistry. 113 (3): 674–82. PMID 20132468. doi:10.1111/j.1471-4159.2010.06631.x.
- ↑ Andreazza AC, Shao L, Wang JF, Young LT (April 2010). "Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder". Archives of General Psychiatry. 67 (4): 360–8. PMID 20368511. doi:10.1001/archgenpsychiatry.2010.22.
- ↑ Moran M, Rivera H, Sánchez-Aragó M, Blázquez A, Merinero B, Ugalde C, Arenas J, Cuezva JM, Martín MA (May 2010). "Mitochondrial bioenergetics and dynamics interplay in complex I-deficient fibroblasts". Biochimica et Biophysica Acta. 1802 (5): 443–53. PMID 20153825. doi:10.1016/j.bbadis.2010.02.001.
- ↑ Binukumar BK, Bal A, Kandimalla R, Sunkaria A, Gill KD (April 2010). "Mitochondrial energy metabolism impairment and liver dysfunction following chronic exposure to dichlorvos". Toxicology. 270 (2–3): 77–84. PMID 20132858. doi:10.1016/j.tox.2010.01.017.
External links
- IST Austria: Sazanov Group MRC MBU Sazanov group
- Interactive Molecular model of NADH dehydrogenase (Requires MDL Chime)
- Complex I homepage
- Electron Transport Complex I at the US National Library of Medicine Medical Subject Headings (MeSH)
As of this edit, this article uses content from "3.D.1 The H+ or Na+-translocating NADH Dehydrogenase (NDH) Family", which is licensed in a way that permits reuse under the Creative Commons Attribution-ShareAlike 3.0 Unported License, but not under the GFDL. All relevant terms must be followed..