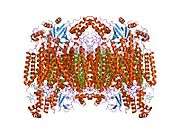
Cytochrome c oxidase subunit I
| Cytochrome c oxidase subunit I | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of the 13-subunit oxidized cytochrome c oxidase.[3] | |||||||||
| Identifiers | |||||||||
| Symbol | COX1 or COI | ||||||||
| Pfam | PF00115 | ||||||||
| InterPro | IPR000883 | ||||||||
| PROSITE | PDOC00074 | ||||||||
| SCOP | 1occ | ||||||||
| SUPERFAMILY | 1occ | ||||||||
| TCDB | 3.D.4 | ||||||||
| OPM superfamily | 4 | ||||||||
| OPM protein | 1v55 | ||||||||
| CDD | cd01663 | ||||||||
| |||||||||

- "Cox1" redirects here. Particularly in a medical context, this can also refer to cyclooxygenase-1 (COX-1).
Cytochrome c oxidase I (COX1) also known as mitochondrially encoded cytochrome c oxidase I (MT-CO1) is a protein that in humans is encoded by the MT-CO1 gene.[4] In other eukaryotes, the gene is called COX1, CO1, or COI.[5] Cytochrome c oxidase I is the main subunit of the cytochrome c oxidase complex.
Function
Cytochrome c oxidase subunit I (CO1 or MT-CO1) is one of three mitochondrial DNA (mtDNA) encoded subunits (MT-CO1, MT-CO2, MT-CO3) of respiratory complex IV. Complex IV is the third and final enzyme of the electron transport chain of mitochondrial oxidative phosphorylation.[4]
Cytochrome c oxidase (EC 1.9.3.1) is a key enzyme in aerobic metabolism. Proton pumping heme-copper oxidases represent the terminal, energy-transfer enzymes of respiratory chains in prokaryotes and eukaryotes. The CuB-heme a3 (or heme o) binuclear centre, associated with the largest subunit I of cytochrome c and ubiquinol oxidases (EC 1.10.3.10), is directly involved in the coupling between dioxygen reduction and proton pumping.[6][7] Some terminal oxidases generate a transmembrane proton gradient across the plasma membrane (prokaryotes) or the mitochondrial inner membrane (eukaryotes).
The enzyme complex consists of 3-4 subunits (prokaryotes) up to 13 polypeptides (mammals) of which only the catalytic subunit (equivalent to mammalian subunit I (COI)) is found in all heme-copper respiratory oxidases. The presence of a bimetallic centre (formed by a high-spin heme and copper B) as well as a low-spin heme, both ligated to six conserved histidine residues near the outer side of four transmembrane spans within COI is common to all family members.[8][9][10] In contrast to eukaryotes the respiratory chain of prokaryotes is branched to multiple terminal oxidases. The enzyme complexes vary in heme and copper composition, substrate type and substrate affinity. The different respiratory oxidases allow the cells to customize their respiratory systems according to a variety of environmental growth conditions.[6]
It has been shown that eubacterial quinol oxidase was derived from cytochrome c oxidase in Gram-positive bacteria and that archaebacterial quinol oxidase has an independent origin. A considerable amount of evidence suggests that proteobacteria (Purple bacteria) acquired quinol oxidase through a lateral gene transfer from Gram-positive bacteria.[6]
A related nitric-oxide reductase (EC 1.7.99.7) exists in denitrifying species of archaea and eubacteria and is a heterodimer of cytochromes b and c. Phenazine methosulphate can act as acceptor. It has been suggested that cytochrome c oxidase catalytic subunits evolved from ancient nitric oxide reductases that could reduce both nitrogen and oxygen.[11][12]
Subfamilies
- Cytochrome c oxidase cbb3-type, subunit I InterPro: IPR004677
- Cytochrome o ubiquinol oxidase, subunit I InterPro: IPR014207
- Cytochrome aa3 quinol oxidase, subunit I InterPro: IPR014233
- Cytochrome c oxidase, subunit I bacterial type InterPro: IPR014241
Application
It is a gene that is often used as a DNA barcode to identify animal species. MT-CO1 gene sequence is suitable for this role because its mutation rate is often fast enough to distinguish closely related species and also because its sequence is conserved among conspecifics. Contrary to the primary objection raised by skeptics that MT-CO1 sequence differences are too small to be detected between closely related species, more than 2% sequence divergence is typically detected between such organisms,[13] suggesting that the barcode is effective. In most if not all seed plants, however, the rate of evolution of cox1 is very slow.
MT-COI (CCOI) in colonic crypts

CCOI is a synonym for MT-COI
CCOI protein is usually expressed at a high level in the cytoplasm of colonic crypts of the human large intestine (colon). However, CCOI is frequently lost in colonic crypts with age in humans and is also often absent in field defects that give rise to colon cancers as well as in portions of colon cancers.[14]
The epithelial inner surface of the colon is punctuated by invaginations, the colonic crypts. The colon crypts are shaped like microscopic thick walled test tubes with a central hole down the length of the tube (the crypt lumen). Four tissue sections are shown in the image in this section, two cut across the long axes of the crypts and two cut parallel to the long axes.
Most of the human colonic crypts in the images have high expression of the brown-orange stained CCOI. However, in some of the colonic crypts all of the cells lack CCOI and appear mostly white, with their main color being the blue-gray staining of the nuclei at the outer walls of the crypts. Greaves et al.[15] showed that deficiencies of CCOI in colonic crypts are due to mutations in the CCOI gene. As seen in panel B, a portion of the stem cells of three crypts appear to have a mutation in CCOI, so that 40% to 50% of the cells arising from those stem cells form a white segment in the cross-cut area.
In humans, the percent of colonic crypts deficient for CCOI is less than 1% before age 40, but then increases linearly with age.[14] On average, the percent of colonic crypts deficient for CCOI reaches 18% in women and 23% in men by 80–84 years of age.[14] Colonic tumors often arise in a field of crypts containing a large cluster (as many as 410) of CCOI-deficient crypts. In colonic cancers, up to 80% of tumor cells can be deficient in CCOI.[14]
As seen in panels C and D, crypts are about 75 to about 110 cells long. The average crypt circumference is 23 cells.[16] Based on these measurements, crypts have between 1725 and 2530 cells. Another report gave a range of 1500 to 4900 cells per colonic crypt.[17]
The occurrence of frequent crypts with almost complete loss of CCOI in their 1700 to 5,000 cells suggests a process of natural selection. However, it has also been shown that a deficiency throughout a particular crypt due to an initial mitochondrial DNA mutation may occasionally occur through a stochastic process.[18][19] Nevertheless, the frequent occurrence of CCOI deficiency in many crypts within a colon epithelium indicates that absence of CCOI likely provides a selective advantage.
CCOI is coded for by the mitochondrial chromosome. There are multiple copies of the chromosome in most mitochondria, usually between 2 and 6 per mitochondrion.[20][21][22] If a mutation occurs in CCOI in one chromosome of a mitochondrion, there may be random segregation of the chromosomes during mitochondrial fission to generate new mitochondria. This can give rise to a mitochondrion with primarily or solely CCOI-mutated chromosomes.
A mitochondrion with largely CCOI-mutated chromosomes would need to have a positive selection bias in order to frequently become the main type of mitochondrion in a cell (a cell with CCOI-deficient homoplasmy). There are about 100 to 700 mitochondria per cell, depending on cell type.[21][22] Furthermore, there is fairly rapid turnover of mitochondria, so that a mitochondrion with CCOI-mutated chromosomes and a positive selection bias could shortly become the major type of mitochondrion in a cell. The average half-life of mitochondria in rats, depending on cell type, is between 9 and 24 days,[23] and in mice is about 2 days.[24] In humans it is likely that the half life of mitochondria is also a matter of days to weeks.
A stem cell at the base of a colonic crypt that was largely CCOI-deficient may compete with the other 4 or 5 stem cells to take over the stem cell niche. If this occurs, then the colonic crypt would be deficient in CCOI in all 1700 to 5,000 cells, as is indicated for some crypts in panels A, B and D of the image.
Crypts of the colon can reproduce by fission, as seen in panel C, where a crypt is fissioning to form two crypts, and in panel B where at least one crypt appears to be fissioning. Most crypts deficient in CCOI are in clusters of crypts (clones of crypts) with two or more CCOI-deficient crypts adjacent to each other (see panel D).[14] This illustrates that clones of deficient crypts often arise, and thus that there is likely a positive selective bias that has allowed them to spread in the human colonic epithelium.
It is not clear why a deficiency of CCOI should have a positive selective bias. One suggestion[14] is that deficiency of CCOI in a mitochondrion leads to lower reactive oxygen production (and less oxidative damage) and this provides a selective advantage in competition with other mitochondria within the same cell to generate homoplasmy for CCOI-deficiency. Another suggestion was that cells with a deficiency in cytochrome c oxidase are apoptosis resistant, and thus more likely to survive. The linkage of CCOI to apoptosis arises because active cytochrome c oxidase oxidizes cytochrome c, which then activates pro-caspase 9, leading to apoptosis.[25] These two factors may contribute to the frequent occurrence of CCOI-deficient colonic crypts with age or during carcinogenesis in the human colon.
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Tsukihara T, Aoyama H, Yamashita E, et al. (May 1996). "The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A". Science. 272 (5265): 1136–44. Bibcode:1996Sci...272.1136T. PMID 8638158. doi:10.1126/science.272.5265.1136.
- 1 2 "Entrez Gene: Cytochrome c oxidase subunit I".
- ↑ Kosakyan A, Heger TJ, Leander BS, Todorov M, Mitchell EA, Lara E (May 2012). "COI barcoding of Nebelid testate amoebae (Amoebozoa: Arcellinida): extensive cryptic diversity and redefinition of the Hyalospheniidae Schultze". Protist. 163 (3): 415–34. PMID 22130576. doi:10.1016/j.protis.2011.10.003.
- 1 2 3 Rumbley J, Gennis RB, Garcia-Horsman JA, Barquera B, Ma J (1994). "The superfamily of heme-copper respiratory oxidases". J. Bacteriol. 176 (18): 5587–5600. PMC 196760
 . PMID 8083153.
. PMID 8083153. - ↑ Glaser P, Villani G, Papa S, Capitanio N (1994). "The proton pump of heme-copper oxidases". Cell Biol. Int. 18 (5): 345–355. PMID 8049679. doi:10.1006/cbir.1994.1084.
- ↑ Saraste M, Castresana J, Higgins DG, Lubben M (1994). "Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen". EMBO J. 13 (11): 2516–2525. PMC 395125
 . PMID 8013452.
. PMID 8013452. - ↑ Capaldi RA, Malatesta F, Darley-Usmar VM (1983). "Structure of cytochrome c oxidase". Biochim. Biophys. Acta. 726 (2): 135–48. PMID 6307356. doi:10.1016/0304-4173(83)90003-4.
- ↑ Saraste M, Holm L, Wikstrom M (1987). "Structural models of the redox centres in cytochrome oxidase". EMBO J. 6 (9): 2819–2823. PMC 553708
 . PMID 2824194.
. PMID 2824194. - ↑ Saraste M, Castresana J (March 1994). "Cytochrome oxidase evolved by tinkering with denitrification enzymes". FEBS Lett. 341 (1): 1–4. PMID 8137905. doi:10.1016/0014-5793(94)80228-9.
- ↑ Chen J, Strous M (February 2013). "Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution". Biochim. Biophys. Acta. 1827 (2): 136–44. PMID 23044391. doi:10.1016/j.bbabio.2012.10.002.
- ↑ Hebert PD, Ratnasingham S, deWaard JR (August 2003). "Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species". Proc. Biol. Sci. 270 Suppl 1: S96–9. PMC 1698023
 . PMID 12952648. doi:10.1098/rsbl.2003.0025.
. PMID 12952648. doi:10.1098/rsbl.2003.0025. - 1 2 3 4 5 6 7 Bernstein C, Facista A, Nguyen H, Zaitlin B, Hassounah N, Loustaunau C, Payne CM, Banerjee B, Goldschmid S, Tsikitis VL, Krouse R, Bernstein H (2010). "Cancer and age related colonic crypt deficiencies in cytochrome c oxidase I". World J Gastrointest Oncol. 2 (12): 429–42. PMC 3011097
 . PMID 21191537. doi:10.4251/wjgo.v2.i12.429.
. PMID 21191537. doi:10.4251/wjgo.v2.i12.429. - ↑ Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, Leedham SJ, Deheragoda M, Sasieni P, Novelli MR, Jankowski JA, Turnbull DM, Wright NA, McDonald SA (2006). "Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission". Proc. Natl. Acad. Sci. U.S.A. 103 (3): 714–9. Bibcode:2006PNAS..103..714G. PMC 1325106
 . PMID 16407113. doi:10.1073/pnas.0505903103.
. PMID 16407113. doi:10.1073/pnas.0505903103. - ↑ Baker AM, Cereser B, Melton S, Fletcher AG, Rodriguez-Justo M, Tadrous PJ, Humphries A, Elia G, McDonald SA, Wright NA, Simons BD, Jansen M, Graham TA (2014). "Quantification of crypt and stem cell evolution in the normal and neoplastic human colon". Cell Rep. 8 (4): 940–7. PMC 4471679
 . PMID 25127143. doi:10.1016/j.celrep.2014.07.019.
. PMID 25127143. doi:10.1016/j.celrep.2014.07.019. - ↑ Nooteboom M, Johnson R, Taylor RW, Wright NA, Lightowlers RN, Kirkwood TB, Mathers JC, Turnbull DM, Greaves LC (2010). "Age-associated mitochondrial DNA mutations lead to small but significant changes in cell proliferation and apoptosis in human colonic crypts". Aging Cell. 9 (1): 96–9. PMC 2816353
 . PMID 19878146. doi:10.1111/j.1474-9726.2009.00531.x.
. PMID 19878146. doi:10.1111/j.1474-9726.2009.00531.x. - ↑ Coller HA, Bodyak ND, Khrapko K (2002). "Frequent intracellular clonal expansions of somatic mtDNA mutations: significance and mechanisms". Ann. N. Y. Acad. Sci. 959: 434–47. Bibcode:2002NYASA.959..434C. PMID 11976216. doi:10.1111/j.1749-6632.2002.tb02113.x.
- ↑ Nekhaeva E, Bodyak ND, Kraytsberg Y, McGrath SB, Van Orsouw NJ, Pluzhnikov A, Wei JY, Vijg J, Khrapko K (2002). "Clonally expanded mtDNA point mutations are abundant in individual cells of human tissues". Proc. Natl. Acad. Sci. U.S.A. 99 (8): 5521–6. Bibcode:2002PNAS...99.5521N. PMC 122802
 . PMID 11943860. doi:10.1073/pnas.072670199.
. PMID 11943860. doi:10.1073/pnas.072670199. - ↑ Legros F, Malka F, Frachon P, Lombès A, Rojo M (2004). "Organization and dynamics of human mitochondrial DNA". J. Cell. Sci. 117 (Pt 13): 2653–62. PMID 15138283. doi:10.1242/jcs.01134.
- 1 2 Robin ED, Wong R (1988). "Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells". J. Cell. Physiol. 136 (3): 507–13. PMID 3170646. doi:10.1002/jcp.1041360316.
- 1 2 Satoh M, Kuroiwa T (1991). "Organization of multiple nucleoids and DNA molecules in mitochondria of a human cell". Exp. Cell Res. 196 (1): 137–40. PMID 1715276. doi:10.1016/0014-4827(91)90467-9.
- ↑ Menzies RA, Gold PH (1971). "The turnover of mitochondria in a variety of tissues of young adult and aged rats". J. Biol. Chem. 246 (8): 2425–9. PMID 5553400.
- ↑ Miwa S, Lawless C, von Zglinicki T (2008). "Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model". Aging Cell. 7 (6): 920–3. PMC 2659384
 . PMID 18691181. doi:10.1111/j.1474-9726.2008.00426.x.
. PMID 18691181. doi:10.1111/j.1474-9726.2008.00426.x. - ↑ Brown GC, Borutaite V (2008). "Regulation of apoptosis by the redox state of cytochrome c". Biochim. Biophys. Acta. 1777 (7–8): 877–81. PMID 18439415. doi:10.1016/j.bbabio.2008.03.024.
Further reading
- Torroni A, Achilli A, Macaulay V, et al. (2006). "Harvesting the fruit of the human mtDNA tree". Trends Genet. 22 (6): 339–45. PMID 16678300. doi:10.1016/j.tig.2006.04.001.
- Bodenteich A, Mitchell LG, Polymeropoulos MH, Merril CR (1993). "Dinucleotide repeat in the human mitochondrial D-loop". Hum. Mol. Genet. 1 (2): 140. PMID 1301157. doi:10.1093/hmg/1.2.140-a.
- Brown MD, Yang CC, Trounce I, et al. (1992). "A mitochondrial DNA variant, identified in Leber hereditary optic neuropathy patients, which extends the amino acid sequence of cytochrome c oxidase subunit I". Am. J. Hum. Genet. 51 (2): 378–85. PMC 1682694
 . PMID 1322638.
. PMID 1322638. - Lu X, Walker T, MacManus JP, Seligy VL (1992). "Differentiation of HT-29 human colonic adenocarcinoma cells correlates with increased expression of mitochondrial RNA: effects of trehalose on cell growth and maturation". Cancer Res. 52 (13): 3718–25. PMID 1377597.
- Marzuki S, Noer AS, Lertrit P, et al. (1992). "Normal variants of human mitochondrial DNA and translation products: the building of a reference data base". Hum. Genet. 88 (2): 139–45. PMID 1757091. doi:10.1007/bf00206061.
- Moraes CT, Andreetta F, Bonilla E, et al. (1991). "Replication-competent human mitochondrial DNA lacking the heavy-strand promoter region". Mol. Cell. Biol. 11 (3): 1631–7. PMC 369459
 . PMID 1996112.
. PMID 1996112. - Attardi G, Chomyn A, Doolittle RF, et al. (1987). "Seven unidentified reading frames of human mitochondrial DNA encode subunits of the respiratory chain NADH dehydrogenase". Cold Spring Harb. Symp. Quant. Biol. 51 (1): 103–14. PMID 3472707. doi:10.1101/sqb.1986.051.01.013.
- Chomyn A, Cleeter MW, Ragan CI, et al. (1986). "URF6, last unidentified reading frame of human mtDNA, codes for an NADH dehydrogenase subunit". Science. 234 (4776): 614–8. Bibcode:1986Sci...234..614C. PMID 3764430. doi:10.1126/science.3764430.
- Chomyn A, Mariottini P, Cleeter MW, et al. (1985). "Six unidentified reading frames of human mitochondrial DNA encode components of the respiratory-chain NADH dehydrogenase". Nature. 314 (6012): 592–7. Bibcode:1985Natur.314..592C. PMID 3921850. doi:10.1038/314592a0.
- Sanger F, Coulson AR, Barrell BG, et al. (1981). "Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing". J. Mol. Biol. 143 (2): 161–78. PMID 6260957. doi:10.1016/0022-2836(80)90196-5.
- Montoya J, Ojala D, Attardi G (1981). "Distinctive features of the 5'-terminal sequences of the human mitochondrial mRNAs". Nature. 290 (5806): 465–70. Bibcode:1981Natur.290..465M. PMID 7219535. doi:10.1038/290465a0.
- Horai S, Hayasaka K, Kondo R, et al. (1995). "Recent African origin of modern humans revealed by complete sequences of hominoid mitochondrial DNAs". Proc. Natl. Acad. Sci. U.S.A. 92 (2): 532–6. Bibcode:1995PNAS...92..532H. PMC 42775
 . PMID 7530363. doi:10.1073/pnas.92.2.532.
. PMID 7530363. doi:10.1073/pnas.92.2.532. - Gattermann N, Retzlaff S, Wang YL, et al. (1997). "Heteroplasmic point mutations of mitochondrial DNA affecting subunit I of cytochrome c oxidase in two patients with acquired idiopathic sideroblastic anemia". Blood. 90 (12): 4961–72. PMID 9389715.
- Bröker S, Meunier B, Rich P, et al. (1998). "MtDNA mutations associated with sideroblastic anaemia cause a defect of mitochondrial cytochrome c oxidase". Eur. J. Biochem. 258 (1): 132–8. PMID 9851701. doi:10.1046/j.1432-1327.1998.2580132.x.
- Andrews RM, Kubacka I, Chinnery PF, et al. (1999). "Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA". Nat. Genet. 23 (2): 147. PMID 10508508. doi:10.1038/13779.
- Karadimas CL, Greenstein P, Sue CM, et al. (2000). "Recurrent myoglobinuria due to a nonsense mutation in the COX I gene of mitochondrial DNA". Neurology. 55 (5): 644–9. PMID 10980727. doi:10.1212/wnl.55.5.644.
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U (2001). "Mitochondrial genome variation and the origin of modern humans". Nature. 408 (6813): 708–13. PMID 11130070. doi:10.1038/35047064.
- Finnilä S, Lehtonen MS, Majamaa K (2001). "Phylogenetic network for European mtDNA". Am. J. Hum. Genet. 68 (6): 1475–84. PMC 1226134
 . PMID 11349229. doi:10.1086/320591.
. PMID 11349229. doi:10.1086/320591. - Maca-Meyer N, González AM, Larruga JM, et al. (2003). "Major genomic mitochondrial lineages delineate early human expansions". BMC Genet. 2: 13. PMC 55343
 . PMID 11553319. doi:10.1186/1471-2156-2-13.
. PMID 11553319. doi:10.1186/1471-2156-2-13.
This article incorporates text from the public domain Pfam and InterPro IPR000883