MMP3
Stromelysin-1 also known as matrix metalloproteinase-3 (MMP-3) is an enzyme that in humans is encoded by the MMP3 gene. The MMP3 gene is part of a cluster of MMP genes which localize to chromosome 11q22.3.[5] MMP-3 has an estimated molecular weight of 54 kDa.[6]
Function
Proteins of the matrix metalloproteinase (MMP) family are involved in the breakdown of extracellular matrix proteins and during tissue remodeling in normal physiological processes, such as embryonic development and reproduction, as well as in disease processes, such as arthritis, and tumour metastasis. Most MMPs are secreted as inactive proproteins which are activated when cleaved by extracellular proteinases.
The MMP-3 enzyme degrades collagen types II, III, IV, IX, and X, proteoglycans, fibronectin, laminin, and elastin. In addition, MMP-3 can also activate other MMPs such as MMP-1, MMP-7, and MMP-9, rendering MMP-3 crucial in connective tissue remodeling.[7] The enzyme is also thought to be involved in wound repair, progression of atherosclerosis, and tumor initiation.
In addition to classical roles for MMP3 in extracellular space, MMP3 can enter in cellular nuclei and control transcription.[8]
Gene regulation
MMP3 itself can enter in nuclei of cells and regulate target gene such as CTGF/CCN2 gene.[8]
Expression of MMP3 is primarily regulated at the level of transcription, where the promoter of the gene responds to various stimuli, including growth factors, cytokines, tumor promoters, and oncogene products.[9] A polymorphism in the promoter of the MMP3 gene was first reported in 1995.[10] The polymorphism is caused by a variation in the number of adenosines located at position -1171 relative to the transcription start site, resulting in one allele having five adenosines (5A) and the other allele having six adenosines (6A). In vitro promoter functional analyses showed that the 5A allele had greater promoter activities as compared with the 6A allele.[7] It has been shown in different studies that individuals carrying the 5A allele have increased susceptibility to diseases attributed to increased MMP expression, such as acute myocardial infarction and abdominal aortic aneurysm.[11][12]
On the other hand, the 6A allele has been found to be associated with diseases characterized by insufficient MMP-3 expression due to a lower promoter activity of the 6A allele, such as progressive coronary atherosclerosis.[7][13][14] The -1171 5A/6A variant has also been associated with congenital anomalies such as cleft lip and palate, where individuals with cleft lip/palate presented significantly more 6A/6A genotypes than controls.[15] Recently, the MMP3 gene was shown to be down-regulated in individuals with cleft lip and palate when compared to controls,[16] reinforcing the nature of cleft lip/palate as a condition resulting from insufficient or defective embryonic tissue remodeling.
Structure

Most members of the MMP family are organized into three basic, distinctive, and well-conserved domains based on structural considerations: an amino-terminal propeptide; a catalytic domain; and a hemopexin-like domain at the carboxy-terminal. The propeptide consists of approximately 80–90 amino acids containing a cysteine residue, which interacts with the catalytic zinc atom via its side chain thiol group. A highly conserved sequence (. . .PRCGXPD. . .) is present in the propeptide. Removal of the propeptide by proteolysis results in zymogen activation, as all members of the MMP family are produced in a latent form.
The catalytic domain contains two zinc ions and at least one calcium ion coordinated to various residues. One of the two zinc ions is present in the active site and is involved in the catalytic processes of the MMPs. The second zinc ion (also known as structural zinc) and the calcium ion are present in the catalytic domain approximately 12 Å away from the catalytic zinc. The catalytic zinc ion is essential for the proteolytic activity of MMPs; the three histidine residues that coordinate with the catalytic zinc are conserved among all the MMPs. Little is known about the roles of the second zinc ion and the calcium ion within the catalytic domain, but the MMPs are shown to possess high affinities for structural zinc and calcium ions.
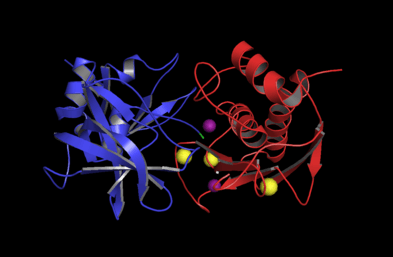
The catalytic domain of MMP-3 can be inhibited by tissue inhibitors of metalloproteinases (TIMPs). The n-terminal fragment of the TIMP binds in the active site cleft much like the peptide substrate would bind. The Cys1 residue of the TIMP chelates to the catalytic zinc and forms hydrogen bonds with one of the carboxylate oxygens of the catalytic glutamate residue (Glu202, see mechanism below). These interactions force the zinc-bound water molecule that is essential to the enzyme's function to leave the enzyme. The loss of the water molecule and the blocking of the active site by TIMP disable the enzyme.[17]
The hemopexin-like domain of MMPs is highly conserved and shows sequence similarity to the plasma protein, hemopexin. The hemopexin-like domain has been shown to play a functional role in substrate binding and/or in interactions with the tissue inhibitors of metalloproteinases (TIMPs), a family of specific MMP protein inhibitors.[18]
Mechanism
The mechanism for MMP-3 is a variation on a larger theme seen in all matrix metalloproteinases. In the active site, a water molecule is coordinated to a glutamate residue (Glu202) and one of the zinc ions present in the catalytic domain. First, the coordinated water molecule performs a nucleophilic attack on the peptide substrate's scissile carbon while the glutamate simultaneously abstracts a proton from the water molecule. The abstracted proton is then removed from the glutamate by the nitrogen of the scissile amide. This forms a tetrahedral gem-diolate intermediate that is coordinated to the zinc atom.[19] In order for the amide product to be released from the active site, the scissile amide must abstract a second proton from the coordinated water molecule.[20] Alternatively, it has been shown for thermolysin (another metalloproteinase) that the amide product can be released in its neutral (R-NH2) form.[21][22] The carboxylate product is released after a water molecule attacks the zinc ion and displaces the carboxylate product.[23] The release of the carboxylate product is thought to be the rate-limiting step in the reaction.[22]
In addition to the water molecule directly involved in the mechanism, a second water molecule is suggested to be a part of the MMP-3 active site. This auxiliary water molecule is thought to stabilize the gem-diolate intermediate as well as the transition states by lowering the activation energy for their formation.[19][24] This is demonstrated in the mechanism and reaction coordinate diagram below.

Disease relevance
MMP-3 has been implicated in exacerbating the effects of traumatic brain injury (TBI) through its disruption of the blood-brain barrier (BBB). Different studies have shown that after the brain undergoes trauma and inflammation has begun, MMP production in the brain is increased.[25][26] In a study conducted using MMP-3 wild type (WT) and knockout (KO) mice, MMP-3 was shown to increase BBB permeability after traumatic injury.[27] The WT mice were shown to have lower claudin-5 and occludin levels than the KO mice after TBI. Claudin and occludin are proteins that are essential for the formation of the tight junctions between the cells of the blood-brain barrier.[28][29] Tissue from uninjured WT and KO mice brains was also treated with active MMP-3. Both the WT and KO tissues showed a drop in claudin-5, occludin, and laminin-α1 (a basal lamina protein), suggesting that MMP-3 directly destroys tight junction and basal lamina proteins.
MMP-3 also does damage to the blood-spinal cord barrier (BSCB), the functional equivalent of the blood-brain barrier,[30] after spinal cord injury (SCI). In a similar study conducted using MMP-3 WT and KO mice, MMP-3 was shown to increase BSCB permeability, with the WT mice showing greater BSCB permeability than the KO mice after spinal cord injury. The same study also found decreased BSCB permeability when spinal cord tissues were treated with a MMP-3 inhibitor. These results suggest that the presence of MMP-3 serves to increase BSCB permeability after SCI.[31] The study showed that MMP-3 accomplishes this damage by degrading claudin-5, occludin, and ZO-1 (another tight junction protein), similar to how MMP-3 damages the BBB.
The increase in blood-brain barrier and blood-spinal cord barrier permeability allows for more neutrophils to infiltrate the brain and spinal cord at the site of inflammation.[27] Neutrophils carry MMP-9.,[32] which has also been shown to degrade occludin.[33] This leads to further disruption of the BBB and BSCB[34]
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000149968 - Ensembl, May 2017
- 1 2 3 GRCm38: Ensembl release 89: ENSMUSG00000043613 - Ensembl, May 2017
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "Entrez Gene: MMP3 matrix metallopeptidase 3 (stromelysin 1, progelatinase)".
- ↑ "Anti-MMP-3 antibody".
- 1 2 3 Ye S, Eriksson P, Hamsten A, Kurkinen M, Humphries SE, Henney AM (May 1996). "Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression". The Journal of Biological Chemistry. 271 (22): 13055–60. PMID 8662692. doi:10.1074/jbc.271.22.13055.
- 1 2 Eguchi T, Kubota S, Kawata K, Mukudai Y, Uehara J, Ohgawara T, Ibaragi S, Sasaki A, Kuboki T, Takigawa M (Apr 2008). "Novel transcription-factor-like function of human matrix metalloproteinase 3 regulating the CTGF/CCN2 gene" (PDF). Molecular and Cellular Biology. 28 (7): 2391–413. PMC 2268440
 . PMID 18172013. doi:10.1128/MCB.01288-07.
. PMID 18172013. doi:10.1128/MCB.01288-07. - ↑ Matrisian LM (Apr 1990). "Metalloproteinases and their inhibitors in matrix remodeling". Trends in Genetics. 6 (4): 121–5. PMID 2132731. doi:10.1016/0168-9525(90)90126-Q.
- ↑ Ye S, Watts GF, Mandalia S, Humphries SE, Henney AM (Mar 1995). "Preliminary report: genetic variation in the human stromelysin promoter is associated with progression of coronary atherosclerosis". British Heart Journal. 73 (3): 209–15. PMC 483800
 . PMID 7727178. doi:10.1136/hrt.73.3.209.
. PMID 7727178. doi:10.1136/hrt.73.3.209. - ↑ Terashima M, Akita H, Kanazawa K, Inoue N, Yamada S, Ito K, Matsuda Y, Takai E, Iwai C, Kurogane H, Yoshida Y, Yokoyama M (Jun 1999). "Stromelysin promoter 5A/6A polymorphism is associated with acute myocardial infarction". Circulation. 99 (21): 2717–9. PMID 10351963. doi:10.1161/01.cir.99.21.2717.
- ↑ Yoon S, Tromp G, Vongpunsawad S, Ronkainen A, Juvonen T, Kuivaniemi H (Nov 1999). "Genetic analysis of MMP3, MMP9, and PAI-1 in Finnish patients with abdominal aortic or intracranial aneurysms". Biochemical and Biophysical Research Communications. 265 (2): 563–8. PMID 10558909. doi:10.1006/bbrc.1999.1721.
- ↑ Humphries SE, Luong LA, Talmud PJ, Frick MH, Kesäniemi YA, Pasternack A, Taskinen MR, Syvänne M (Jul 1998). "The 5A/6A polymorphism in the promoter of the stromelysin-1 (MMP-3) gene predicts progression of angiographically determined coronary artery disease in men in the LOCAT gemfibrozil study. Lopid Coronary Angiography Trial". Atherosclerosis. 139 (1): 49–56. PMID 9699891. doi:10.1016/S0021-9150(98)00053-7.
- ↑ de Maat MP, Jukema JW, Ye S, Zwinderman AH, Moghaddam PH, Beekman M, Kastelein JJ, van Boven AJ, Bruschke AV, Humphries SE, Kluft C, Henney AM (Mar 1999). "Effect of the stromelysin-1 promoter on efficacy of pravastatin in coronary atherosclerosis and restenosis". The American Journal of Cardiology. 83 (6): 852–6. PMID 10190398. doi:10.1016/S0002-9149(98)01073-X.
- ↑ Letra A, Silva RA, Menezes R, Astolfi CM, Shinohara A, de Souza AP, Granjeiro JM (Oct 2007). "MMP gene polymorphisms as contributors for cleft lip/palate: association with MMP3 but not MMP1". Archives of Oral Biology. 52 (10): 954–60. PMID 17537400. doi:10.1016/j.archoralbio.2007.04.005.
- ↑ Bueno DF, Sunaga DY, Kobayashi GS, Aguena M, Raposo-Amaral CE, Masotti C, Cruz LA, Pearson PL, Passos-Bueno MR (Jun 2011). "Human stem cell cultures from cleft lip/palate patients show enrichment of transcripts involved in extracellular matrix modeling by comparison to controls". Stem Cell Reviews. 7 (2): 446–57. PMC 3073041
 . PMID 21052871. doi:10.1007/s12015-010-9197-3.
. PMID 21052871. doi:10.1007/s12015-010-9197-3. - ↑ Gomis-Rüth FX, Maskos K, Betz M, Bergner A, Huber R, Suzuki K, Yoshida N, Nagase H, Brew K, Bourenkov GP, Bartunik H, Bode W (Sep 1997). "Mechanism of inhibition of the human matrix metalloproteinase stromelysin-1 by TIMP-1". Nature. 389 (6646): 77–81. PMID 9288970. doi:10.1038/37995.
- ↑ Massova I, Kotra LP, Fridman R, Mobashery S (Sep 1998). "Matrix metalloproteinases: structures, evolution, and diversification". FASEB Journal. 12 (12): 1075–95. PMID 9737711. doi:10.1142/S0217984998001256.
- 1 2 Pelmenschikov V, Siegbahn PE (Nov 2002). "Catalytic mechanism of matrix metalloproteinases: two-layered ONIOM study". Inorganic Chemistry. 41 (22): 5659–66. PMID 12401069. doi:10.1021/ic0255656.
- ↑ Hangauer DG, Monzingo AF, Matthews BW (Nov 1984). "An interactive computer graphics study of thermolysin-catalyzed peptide cleavage and inhibition by N-carboxymethyl dipeptides". Biochemistry. 23 (24): 5730–41. PMID 6525336. doi:10.1021/bi00319a011.
- ↑ Pelmenschikov V, Blomberg MR, Siegbahn PE (Mar 2002). "A theoretical study of the mechanism for peptide hydrolysis by thermolysin". Journal of Biological Inorganic Chemistry. 7 (3): 284–98. PMID 11935352. doi:10.1007/s007750100295.
- 1 2 Vasilevskaya T, Khrenova MG, Nemukhin AV, Thiel W (Aug 2015). "Mechanism of proteolysis in matrix metalloproteinase-2 revealed by QM/MM modeling". Journal of Computational Chemistry. 36 (21): 1621–30. PMID 26132652. doi:10.1002/jcc.23977.
- ↑ Harrison RK, Chang B, Niedzwiecki L, Stein RL (Nov 1992). "Mechanistic studies on the human matrix metalloproteinase stromelysin". Biochemistry. 31 (44): 10757–62. PMID 1420192. doi:10.1021/bi00159a016.
- ↑ Browner MF, Smith WW, Castelhano AL (May 1995). "Matrilysin-inhibitor complexes: common themes among metalloproteases". Biochemistry. 34 (20): 6602–10. PMID 7756291. doi:10.1021/bi00020a004.
- ↑ Falo MC, Fillmore HL, Reeves TM, Phillips LL (Sep 2006). "Matrix metalloproteinase-3 expression profile differentiates adaptive and maladaptive synaptic plasticity induced by traumatic brain injury". Journal of Neuroscience Research. 84 (4): 768–81. PMID 16862547. doi:10.1002/jnr.20986.
- ↑ Morita-Fujimura Y, Fujimura M, Gasche Y, Copin JC, Chan PH (Jan 2000). "Overexpression of copper and zinc superoxide dismutase in transgenic mice prevents the induction and activation of matrix metalloproteinases after cold injury-induced brain trauma". Journal of Cerebral Blood Flow and Metabolism. 20 (1): 130–8. PMID 10616801. doi:10.1097/00004647-200001000-00017.
- 1 2 Gurney KJ, Estrada EY, Rosenberg GA (Jul 2006). "Blood-brain barrier disruption by stromelysin-1 facilitates neutrophil infiltration in neuroinflammation". Neurobiology of Disease. 23 (1): 87–96. PMID 16624562. doi:10.1016/j.nbd.2006.02.006.
- ↑ Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S (Jun 1998). "Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin". The Journal of Cell Biology. 141 (7): 1539–50. PMC 2132999
 . PMID 9647647. doi:10.1083/jcb.141.7.1539.
. PMID 9647647. doi:10.1083/jcb.141.7.1539. - ↑ Nitta T, Hata M, Gotoh S, Seo Y, Sasaki H, Hashimoto N, Furuse M, Tsukita S (May 2003). "Size-selective loosening of the blood-brain barrier in claudin-5-deficient mice". The Journal of Cell Biology. 161 (3): 653–60. PMC 2172943
 . PMID 12743111. doi:10.1083/jcb.200302070.
. PMID 12743111. doi:10.1083/jcb.200302070. - ↑ Bartanusz V, Jezova D, Alajajian B, Digicaylioglu M (Aug 2011). "The blood-spinal cord barrier: morphology and clinical implications". Annals of Neurology. 70 (2): 194–206. PMID 21674586. doi:10.1002/ana.22421.
- ↑ Lee JY, Choi HY, Ahn HJ, Ju BG, Yune TY (Nov 2014). "Matrix metalloproteinase-3 promotes early blood-spinal cord barrier disruption and hemorrhage and impairs long-term neurological recovery after spinal cord injury". The American Journal of Pathology. 184 (11): 2985–3000. PMID 25325922. doi:10.1016/j.ajpath.2014.07.016.
- ↑ Opdenakker G, Van den Steen PE, Dubois B, Nelissen I, Van Coillie E, Masure S, Proost P, Van Damme J (Jun 2001). "Gelatinase B functions as regulator and effector in leukocyte biology". Journal of Leukocyte Biology. 69 (6): 851–9. PMID 11404367.
- ↑ Giebel SJ, Menicucci G, McGuire PG, Das A (May 2005). "Matrix metalloproteinases in early diabetic retinopathy and their role in alteration of the blood-retinal barrier". Laboratory Investigation; A Journal of Technical Methods and Pathology. 85 (5): 597–607. PMID 15711567. doi:10.1038/labinvest.3700251.
- ↑ Aubé B, Lévesque SA, Paré A, Chamma É, Kébir H, Gorina R, Lécuyer MA, Alvarez JI, De Koninck Y, Engelhardt B, Prat A, Côté D, Lacroix S (Sep 2014). "Neutrophils mediate blood-spinal cord barrier disruption in demyelinating neuroinflammatory diseases". Journal of Immunology. 193 (5): 2438–54. PMID 25049355. doi:10.4049/jimmunol.1400401.
Further reading
- Matrisian LM (Apr 1990). "Metalloproteinases and their inhibitors in matrix remodeling". Trends in Genetics. 6 (4): 121–5. PMID 2132731. doi:10.1016/0168-9525(90)90126-Q.
- Massova I, Kotra LP, Fridman R, Mobashery S (Sep 1998). "Matrix metalloproteinases: structures, evolution, and diversification". FASEB Journal. 12 (12): 1075–95. PMID 9737711. doi:10.1142/S0217984998001256.
- Nagase H, Woessner JF (Jul 1999). "Matrix metalloproteinases". The Journal of Biological Chemistry. 274 (31): 21491–4. PMID 10419448. doi:10.1074/jbc.274.31.21491.
- Lijnen HR (Jan 2002). "Matrix metalloproteinases and cellular fibrinolytic activity". Biochemistry. Biokhimii︠a︡. 67 (1): 92–8. PMID 11841344. doi:10.1023/A:1013908332232.