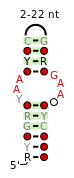
Internal loop
Internal-loops (also termed interior loops) in RNA are found where the double stranded RNA separates due to no Watson-Crick base pairing between the nucleotides. Internal-loops differ from Stem-loops as they occur in middle of a stretch of double stranded RNA. The non-canonicoal residues result in the double helix becoming distorted due to unwinding, unstacking and kinking.
Internal-loops can be classified as either symmetrical or asymmetrical, with some asymmetrical internal-loops, also known as bulges. Many important structural motifs are composed of internal loops such as the C-loop,[1] the docking-elbow,[2] kink-turns (k-turn),[3][4] the right-angle,[5] the sarcin/ricin loops (also called bulged-G motifs),[6][7][8] the twist-up motif[9] and the UAA/GAN internal loop motif.[10]
References
- ↑ Lescoute, A; Leontis, NB; Massire, C; Westhof, E (2005). "Recurrent structural RNA motifs, Isostericity Matrices and sequence alignments.". Nucleic Acids Research. 33 (8): 2395–409. PMC 1087784
 . PMID 15860776. doi:10.1093/nar/gki535.
. PMID 15860776. doi:10.1093/nar/gki535. - ↑ Lehmann, J; Jossinet, F; Gautheret, D (May 1, 2013). "A universal RNA structural motif docking the elbow of tRNA in the ribosome, RNAse P and T-box leaders.". Nucleic Acids Research. 41 (10): 5494–502. PMC 3664808
 . PMID 23580544. doi:10.1093/nar/gkt219.
. PMID 23580544. doi:10.1093/nar/gkt219. - ↑ Klein, D.J. (2001). "The kink-turn: a new RNA secondary structure motif". The EMBO Journal. 20 (15): 4214–4221. ISSN 1460-2075. doi:10.1093/emboj/20.15.4214.
- ↑ Schroeder, KT; McPhee, SA; Ouellet, J; Lilley, DM (Aug 2010). "A structural database for k-turn motifs in RNA.". RNA (New York, N.Y.). 16 (8): 1463–8. PMC 2905746
 . PMID 20562215. doi:10.1261/rna.2207910.
. PMID 20562215. doi:10.1261/rna.2207910. - ↑ Grabow, WW; Zhuang, Z; Swank, ZN; Shea, JE; Jaeger, L (Nov 23, 2012). "The right angle (RA) motif: a prevalent ribosomal RNA structural pattern found in group I introns.". Journal of Molecular Biology. 424 (1–2): 54–67. PMC 3488136
 . PMID 22999957. doi:10.1016/j.jmb.2012.09.012.
. PMID 22999957. doi:10.1016/j.jmb.2012.09.012. - 1 2 3 Szewczak, AA; Moore, PB (Mar 17, 1995). "The sarcin/ricin loop, a modular RNA.". Journal of Molecular Biology. 247 (1): 81–98. PMID 7897662. doi:10.1006/jmbi.1994.0124.
- ↑ Leontis, NB; Westhof, E (Oct 30, 1998). "A common motif organizes the structure of multi-helix loops in 16 S and 23 S ribosomal RNAs.". Journal of Molecular Biology. 283 (3): 571–83. PMID 9784367. doi:10.1006/jmbi.1998.2106.
- ↑ Moore PB (1999). "Structural motifs in RNA". Annu. Rev. Biochem. 68: 287–300. PMID 10872451. doi:10.1146/annurev.biochem.68.1.287.
- 1 2 Zhong, C; Zhang, S (Feb 2012). "Clustering RNA structural motifs in ribosomal RNAs using secondary structural alignment.". Nucleic Acids Research. 40 (3): 1307–17. PMC 3273805
 . PMID 21976732. doi:10.1093/nar/gkr804.
. PMID 21976732. doi:10.1093/nar/gkr804. - 1 2 Lee, JC; Gutell, RR; Russell, R (Jul 28, 2006). "The UAA/GAN internal loop motif: a new RNA structural element that forms a cross-strand AAA stack and long-range tertiary interactions.". Journal of Molecular Biology. 360 (5): 978–88. PMID 16828489. doi:10.1016/j.jmb.2006.05.066.