Immunoglobulin heavy chain
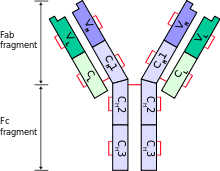

The immunoglobulin heavy chain (IgH) is the large polypeptide subunit of an antibody (immunoglobulin).
A typical antibody is composed of two immunoglobulin (Ig) heavy chains and two Ig light chains. Several different types of heavy chain exist that define the class or isotype of an antibody. These heavy chain types vary between different animals. All heavy chains contain a series of immunoglobulin domains, usually with one variable domain (VH) that is important for binding antigen and several constant domains (CH1, CH2, etc.). Production of the a viable heavy chain is a key step in B cell maturation. If the heavy chain is able to bind to a surrogate light chain and move to the plasma membrane, then the developing B cell can begin producing its light chain.[2]
The heavy chain doesn't always have to bind to a light chain. Pre-B lymphocytes can synthesize heavy chain in the absence of light chain, which then can allow the heavy chain to bind to a heavy-chain binding protein.[3]
In mammals
Classes
There are five types of mammalian immunoglobulin heavy chain: γ, δ, α, μ and ε.[4] They define classes of immunoglobulins: IgG, IgD, IgA, IgM and IgE, respectively.
- Heavy chains α and γ have approximately 450 amino acids.
- Heavy chains μ and ε have approximately 550 amino acids.[4]
Regions
Each heavy chain has two regions:
- a constant region (which is the same for all immunoglobulins of the same class but differs between classes).
- a variable region that differs between different B cells, but is the same for all immunoglobulins produced by the same B cell or B cell clone. The variable domain of any heavy chain is composed of a single immunoglobulin domain. These domains are about 110 amino acids long.
Cows
Cows, specifically Bos Taurus, show a variation on the general mammalian theme in which the heavy chain CDR H3 region has adapted to produce a divergent repertoire of antibodies which present a "stalk and knob" antigen interaction surface instead of the more familiar bivalent tip surface.[6] The bovine CDR is unusually long and contains unique sequence attributes which support the production of paired cysteine residues during somatic hypermutation.[6] Thus, where in humans the somatic hypermutation step targets the V(D)J recombination process, the target in cows is on the creation of diverse disulfide bonds and the generation of unique sets of loops which interact with antigen.[6] A speculated evolutionary driver for this variation is the presence of a vastly more diverse microbial environment in the digestive system of the cow as a consequence of their being ruminants.[6]
In fish
Jawed fish appear to be the most primitive animals that are able to make antibodies like those described for mammals.[7] However, fish do not have the same repertoire of antibodies that mammals possess.[8] Three distinct Ig heavy chains have so far been identified in bony fish.
- The first identified was the μ (or mu) heavy chain that is present in all jawed fish and is the heavy chain for what is thought to be the primordial immunoglobulin. The resulting antibody, IgM, is secreted as a tetramer in teleost fish instead of the typical pentamer found in mammals and sharks.
- The heavy chain (δ) for IgD was identified initially from the channel catfish and Atlantic salmon and is now well documented for many teleost fish.[9]
- The third teleost Ig heavy chain gene was identified very recently and does not resemble any of the heavy chains so far described for mammals. This heavy chain, identified in both rainbow trout (τ)[10] and zebrafish (ζ),[11] could potentially form a distinct antibody isotype (IgT or IgZ) that may precede IgM in evolutionary terms.
Similar to the situation observed for bony fish, three distinct Ig heavy chain isotypes have been identified in cartilaginous fish. With the exception of μ, these Ig heavy chain isotypes appear to be unique to cartilaginous fish. The resulting antibodies are designated IgW (also called IgX or IgNARC) and IgNAR ('immunoglobulin new antigen receptor').[12][13] The latter type is a heavy-chain antibody, an antibody lacking light chains, and can be used to produce single-domain antibodies, which are essentially the variable domain of an IgNAR.[14]
IgW has now also been found in the group of lobe finned fishes including the coelacanth and lungfish. The IgW1 and IgW2 in coelacanth has a usual (VD)n-Jn-C structure as well has having a large number of constant domains.[15][16]
In amphibians
Frogs can synthesize IgX and IgY. [17]
See also
References
- ↑ "Archived copy". Archived from the original on April 19, 2007. Retrieved April 20, 2007.
- ↑ Mårtensson, I-L; Ceredig, R (2017-01-23). "Role of the surrogate light chain and the pre-B-cell receptor in mouse B-cell development". Immunology. 101 (4): 435–441. ISSN 0019-2805. PMC 2327112
 . PMID 11122446. doi:10.1046/j.1365-2567.2000.00151.x.
. PMID 11122446. doi:10.1046/j.1365-2567.2000.00151.x. - ↑ Haas, Ingrid G.; Wabl, Matthias (1983). "Immunoglobulin heavy chain binding protein". Nature. 306 (5941): 387–9. Bibcode:1983Natur.306..387H. PMID 6417546. doi:10.1038/306387a0.
- 1 2 3 Janeway CA, Jr.; et al. (2001). Immunobiology (5th ed.). Garland Publishing. ISBN 0-8153-3642-X. (electronic full text via NCBI Bookshelf).
- ↑ Woof, Jenny M.; Burton, Dennis R. (2004). "Human antibody–Fc receptor interactions illuminated by crystal structures". Nature Reviews Immunology. 4 (2): 89–99. PMID 15040582. doi:10.1038/nri1266.
- 1 2 3 4 Wang, Feng; Ekiert, Damian C.; Ahmad, Insha; Yu, Wenli; Zhang, Yong; Bazirgan, Omar; Torkamani, Ali; Raudsepp, Terje; Mwangi, Waithaka; Criscitiello, Michael F.; Wilson, Ian A.; Schultz, Peter G.; Smider, Vaughn V. (2013). "Reshaping Antibody Diversity". Cell. 153 (6): 1379–93. PMC 4007204
 . PMID 23746848. doi:10.1016/j.cell.2013.04.049.
. PMID 23746848. doi:10.1016/j.cell.2013.04.049. - ↑ Fish heavy chain and light chain genes Archived March 23, 2007, at the Wayback Machine.
- ↑ Bengtén, Eva; Clem, L. William; Miller, Norman W.; Warr, Gregory W.; Wilson, Melanie (2006). "Channel catfish immunoglobulins: Repertoire and expression". Developmental & Comparative Immunology. 30: 77. doi:10.1016/j.dci.2005.06.016.
- ↑ Solem, Stein Tore; Stenvik, Jørgen (2006). "Antibody repertoire development in teleosts—a review with emphasis on salmonids and Gadus morhua L". Developmental & Comparative Immunology. 30: 57. doi:10.1016/j.dci.2005.06.007.
- ↑ Hansen, J. D.; Landis, E. D.; Phillips, R. B. (2005). "Discovery of a unique Ig heavy-chain isotype (IgT) in rainbow trout: Implications for a distinctive B cell developmental pathway in teleost fish". Proceedings of the National Academy of Sciences. 102 (19): 6919–6924. Bibcode:2005PNAS..102.6919H. JSTOR 3375456. PMC 1100771
 . PMID 15863615. doi:10.1073/pnas.0500027102.
. PMID 15863615. doi:10.1073/pnas.0500027102. - ↑ Danilova, Nadia; Bussmann, Jeroen; Jekosch, Kerstin; Steiner, Lisa A (2005). "The immunoglobulin heavy-chain locus in zebrafish: Identification and expression of a previously unknown isotype, immunoglobulin Z". Nature Immunology. 6 (3): 295–302. PMID 15685175. doi:10.1038/ni1166.
- ↑ Dooley, H.; Flajnik, M.F. (2006). "Antibody repertoire development in cartilaginous fish". Developmental & Comparative Immunology. 30: 43. doi:10.1016/j.dci.2005.06.022.
- ↑ Simmons, David P.; Abregu, Fiona A.; Krishnan, Usha V.; Proll, David F.; Streltsov, Victor A.; Doughty, Larissa; Hattarki, Meghan K.; Nuttall, Stewart D. (2006). "Dimerisation strategies for shark IgNAR single domain antibody fragments". Journal of Immunological Methods. 315 (1–2): 171–84. PMID 16962608. doi:10.1016/j.jim.2006.07.019.
- ↑ Wesolowski, Janusz; Alzogaray, Vanina; Reyelt, Jan; Unger, Mandy; Juarez, Karla; Urrutia, Mariela; Cauerhff, Ana; Danquah, Welbeck; Rissiek, Björn; Scheuplein, Felix; Schwarz, Nicole; Adriouch, Sahil; Boyer, Olivier; Seman, Michel; Licea, Alexei; Serreze, David V.; Goldbaum, Fernando A.; Haag, Friedrich; Koch-Nolte, Friedrich (2009). "Single domain antibodies: Promising experimental and therapeutic tools in infection and immunity". Medical Microbiology and Immunology. 198 (3): 157–74. PMC 2714450
 . PMID 19529959. doi:10.1007/s00430-009-0116-7.
. PMID 19529959. doi:10.1007/s00430-009-0116-7. - ↑ Zhang, Tianyi; Tacchi, Luca; Wei, Zhiguo; Zhao, Yaofeng; Salinas, Irene (2014). "Intraclass diversification of immunoglobulin heavy chain genes in the African lungfish". Immunogenetics. 66 (5): 335–51. PMC 4348116
 . PMID 24676685. doi:10.1007/s00251-014-0769-2.
. PMID 24676685. doi:10.1007/s00251-014-0769-2. - ↑ Ota, T.; Rast, J. P.; Litman, G. W.; Amemiya, C. T. (2003). "Lineage-restricted retention of a primitive immunoglobulin heavy chain isotype within the Dipnoi reveals an evolutionary paradox". Proceedings of the National Academy of Sciences. 100 (5): 2501–6. PMC 151370
 . PMID 12606718. doi:10.1073/pnas.0538029100.
. PMID 12606718. doi:10.1073/pnas.0538029100. - ↑ Du, Christina C.; Mashoof, Sara M.; Criscitiello, Michael F. (2012). "Oral immunization of the African clawed frog (Xenopus laevis) upregulates the mucosal immunoglobulin IgX". Veterinary Immunology and Immunopathology. 145 (1–2): 493–8. PMC 3273591
 . PMID 22100190. doi:10.1016/j.vetimm.2011.10.019.
. PMID 22100190. doi:10.1016/j.vetimm.2011.10.019.
External links
- Immunoglobulin Heavy Chains at the US National Library of Medicine Medical Subject Headings (MeSH)
- Educational Resource for Heavy Chain Analysis