Mass spectrometry imaging
Mass spectrometry imaging (MSI) (also known as imaging mass spectrometry) is a technique used in mass spectrometry to visualize the spatial distribution of chemical compositions e.g. compounds, biomarkers, metabolites, peptides or proteins by their molecular masses. Although widely used traditional methodologies like radiochemistry and immunohistochemistry achieve the same goal as MSI, they are limited in their abilities to analyze multiple samples at once, and can prove to be lacking if researchers do not have prior knowledge of the samples being studied.[1] Emerging technologies in the field of MSI are MALDI imaging and secondary ion mass spectrometry imaging (SIMS imaging).[2][3]
SIMS imaging
_Imaging.png)
Secondary ion mass spectrometry (SIMS) is used to analyze solid surfaces and thin films by sputtering the surface with a focused primary ion beam and collecting and analyzing ejected secondary ions. There are many different sources for a primary ion beam. However, the primary ion beam must contain ions that are at the higher end of the energy scale. Some common sources are: Cs+, O2+, O, Ar+ and Ga+.[5] SIMS imaging is performed in a manner similar to electron microscopy; the primary ion beam is emitted across the sample while secondary mass spectra are recorded.[6] SIMS proves to be advantageous in providing increased resolution for visualizing spatial distribution over smaller mass ranges.[1] SIMS is widely regarded as one of the most sensitive forms of mass spectrometry as it can detect elements as small as 10−6-10−9.[7]
Multiplexed ion beam imaging (MIBI) is a SIMS method that uses metal isotope labeled antibodies to label compounds in biological samples.[8]
Developments within SIMS: Some chemical modifications have been made within SIMS to increase the efficiency of the process. There are currently two separate techniques being used to help increase the overall efficiency by increasing the sensitivity of SIMS measurements: matrix-enhanced SIMS (ME-SIMS) - This has the same sample preparation as MALDI does as this simulates the chemical ionization properties of MALDI. ME-SIMS does not sample nearly as much material. However, if the analyte being tested has a low mass value then it can produce a similar looking spectra to that of a MALDI spectra. ME-SIMS has been so effective that it has been able to detect low mass chemicals at sub cellular levels that was not possible prior to the development of the ME-SIMS technique.[3] The second technique being used is called sample metallization (Meta-SIMS) - This is the process of gold or silver addition to the sample. This forms a layer of gold or silver around the sample and it is normally no more than 1-3 nm thick. Using this technique has resulted in an increase of sensitivity for larger mass samples. The addition of the metallic layer also allows for the conversion of insulating samples to conducting samples, thus charge compensation within SIMS experiments is no longer required.[9]
MALDI imaging
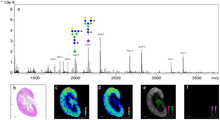
Matrix-assisted laser desorption ionization can be used as a mass spectrometry imaging technique for relatively large molecules.[3] It has recently been shown that the most effective type of matrix to use is an ionic matrix for MALDI imaging of tissue. In this version of the technique the sample, typically a thin tissue section, is moved in two dimensions while the mass spectrum is recorded.[11] Although MALDI has the benefit of being able to record the spatial distribution of larger molecules, it comes at the cost of lower resolution than the SIMS technique. MALDI experiments commonly use either an Nd:YAG (355 nm) or N2 (337 nm) laser for ionization.[3]
Pharmacodynamics and toxicodynamics in tissue have been studied by MALDI imaging.[12]
Ambient ionization methods
Ambient ionization methods such as desorption electrospray ionization (DESI),[13] laser ablation electrospray ionization (LAESI),[14] nanospray desorption electrospray ionization (nano-DESI),[15] matrix-assisted laser desorption electrospray ionization (MALDESI),[16] and liquid extraction surface anaysis (LESA),[17] have also been employed for MSI.
Standard data format for mass spectrometry imaging datasets
As an extension of the mzML standard format developed by HUPO for mass spectrometry datasets, the Computis project developed the imzML standard format for mass spectrometry imaging datasets.[18]
Software
There are many free software packages available for visualization and mining of imaging mass spectrometry data. Converters from Thermo Fisher format, Analyze format, GRD format and Bruker format to imzML format were developed by the Computis project. Some software modules are also available for viewing mass spectrometry images in imzML format: Biomap] (Novartis, free), Datacube Explorer (AMOLF, free),[19] EasyMSI (CEA), Mirion (JLU), MSiReader (NCSU, free)[20] and SpectralAnalysis.[21]
For processing .imzML files with the free statistical and graphics language R, a collection of R scripts is available, which permits parallel-processing of large files on a local computer, a remote cluster or on the Amazon cloud.[22]
Another free statistical package for processing imzML and Analyze 7.5 data in R exists, Cardinal .[23]
References
- 1 2 Monroe E, Annangudi S, Hatcher N, Gutstein H, Rubakhin S, Sweedler J (2008). "SIMS and MADLI MS Imaging of the spinal cord". Proteomics. 8 (18): 3746–3754. doi:10.1002/pmic.200800127.
- ↑ Rohner T, Staab D, Stoeckli M (2005). "MALDI mass spectrometric imaging of biological tissue sections". Mechanisms of Ageing and Development. 126 (1): 177–185. PMID 15610777. doi:10.1016/j.mad.2004.09.032.
- 1 2 3 4 McDonnell LA, Heeren RM (2007). "Imaging mass spectrometry". Mass spectrometry reviews. 26 (4): 606–43. PMID 17471576. doi:10.1002/mas.20124.
- ↑ DeBord J, Smith D, Anderton C, Heeren R, Paša-Tolić L, Gomer R, Fernandez-Lima F. "Secondary ion mass spectrometry imaging of dictyostelium discoideum aggregation streams". PLoS ONE. 9: e99319. PMC 4049834
 . PMID 24911189. doi:10.1371/journal.pone.0099319.
. PMID 24911189. doi:10.1371/journal.pone.0099319. - ↑ Amstalden Van Hove E, Smith D, Heeren R (2010). "A concise review of mass spectrometry imaging". Journal of Chromatography A. 1217 (25): 3946–3954. doi:10.1016/j.chroma.2010.01.033.
- ↑ Penner-Hahn, James E. (2013). "Chapter 2. Technologies for Detecting Metals in Single Cells. Section 2.1, Secondary Ion Mass Specctrometry". In Banci, Lucia (Ed.). Metallomics and the Cell. Metal Ions in Life Sciences. 12. Springer. ISBN 978-94-007-5560-4. doi:10.1007/978-94-007-5561-1_2.electronic-book ISBN 978-94-007-5561-1 ISSN 1559-0836electronic-ISSN 1868-0402
- ↑ Chabala J, Soni K, Li J, Gavlirov K, Levi-Setti R (1995). "High resolution chemical imaging with scanning ion probe SIMS": 191–212.
- ↑ Angelo, Michael; Bendall, Sean C; Finck, Rachel; Hale, Matthew B; Hitzman, Chuck; Borowsky, Alexander D; Levenson, Richard M; Lowe, John B; Liu, Scot D; Zhao, Shuchun; Natkunam, Yasodha; Nolan, Garry P (2014). "Multiplexed ion beam imaging of human breast tumors". Nature Medicine. 20 (4): 436–442. ISSN 1078-8956. PMC 4110905
 . PMID 24584119. doi:10.1038/nm.3488.
. PMID 24584119. doi:10.1038/nm.3488. - ↑ Delcorte A, Befahy S, Poleunis C, Troosters M, Bertrand P. "Improvements of metal adhesion to silicon films: a ToF-SIMS study".
- ↑ Powers, Thomas W.; Neely, Benjamin A.; Shao, Yuan; Tang, Huiyuan; Troyer, Dean A.; Mehta, Anand S.; Haab, Brian B.; Drake, Richard R. (2014). "MALDI Imaging Mass Spectrometry Profiling of N-Glycans in Formalin-Fixed Paraffin Embedded Clinical Tissue Blocks and Tissue Microarrays". PLoS ONE. 9 (9): e106255. ISSN 1932-6203. PMC 4153616
 . PMID 25184632. doi:10.1371/journal.pone.0106255.
. PMID 25184632. doi:10.1371/journal.pone.0106255. - ↑ Chaurand P, Norris JL, Cornett DS, Mobley JA, Caprioli RM (2006). "New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry". J. Proteome Res. 5 (11): 2889–900. PMID 17081040. doi:10.1021/pr060346u.
- ↑ Patel, Ekta (1 January 2015). "MALDI-MS imaging for the study of tissue pharmacodynamics and toxicodynamics". Bioanalysis. 7 (1): 91–101. doi:10.4155/bio.14.280. Retrieved 24 February 2015.
- ↑ Ifa DR, Wiseman JM, Song Q, Cooks RG (2007). "Development of capabilities for imaging mass spectrometry under ambient conditions with desorption electrospray ionization (DESI)". International Journal of Mass Spectrometry. 259 (1–3): 8–15. doi:10.1016/j.ijms.2006.08.003.
- ↑ Nemes, Peter; Woods, Amina S.; Vertes, Akos (2010-02-01). "Simultaneous Imaging of Small Metabolites and Lipids in Rat Brain Tissues at Atmospheric Pressure by Laser Ablation Electrospray Ionization Mass Spectrometry". Analytical Chemistry. 82 (3): 982–988. ISSN 0003-2700. PMC 2964874
 . PMID 20050678. doi:10.1021/ac902245p.
. PMID 20050678. doi:10.1021/ac902245p. - ↑ Lanekoff, Ingela; Heath, Brandi S.; Liyu, Andrey; Thomas, Mathew; Carson, James P.; Laskin, Julia (2012-10-02). "Automated Platform for High-Resolution Tissue Imaging Using Nanospray Desorption Electrospray Ionization Mass Spectrometry". Analytical Chemistry. 84 (19): 8351–8356. ISSN 0003-2700. doi:10.1021/ac301909a.
- ↑ Robichaud, G.; Barry, J.A.; Garrard, K. P.; Muddiman, D.C. (2013). "Infrared Matrix-Assisted Laser Desorption Electrospray Ionization (IR-MALDESI) Imaging Source Coupled to a FT-ICR Mass Spectrometer". J. Am. Soc. Mass Spectrom. 24: 92–100. doi:10.1007/s13361-012-0505-9.
- ↑ Griffiths, Rian L.; Creese, Andrew J.; Race, Alan M.; Bunch, Josephine; Cooper, Helen J. (2016-07-05). "LESA FAIMS Mass Spectrometry for the Spatial Profiling of Proteins from Tissue". Analytical Chemistry. 88 (13): 6758–6766. ISSN 0003-2700. doi:10.1021/acs.analchem.6b01060.
- ↑ A. Römpp; T. Schramm; A. Hester; I. Klinkert; J.P. Both; R.M.A. Heeren; M. Stoeckli; B. Spengler (2011). "Chapter imzML: Imaging Mass Spectrometry Markup Language: A Common Data Format for Mass Spectrometry Imaging in Data Mining in Proteomics: From Standards to Applications". Methods in Molecular Biology, Humana Press, New York. 696. pp. 205–224.
- ↑ Klinkert, I.; Chughtai, K.; Ellis, S. R.; Heeren, R. M. A. (2014). "Methods for Full Resolution Data Exploration and Visualization for Large 2D and 3D Mass Spectrometry Imaging Datasets". International Journal of Mass Spectrometry. 362: 40–47. doi:10.1016/j.ijms.2013.12.012.
- ↑ Robichaud, G.; Garrard, K. P.; Barry, J. A.; Muddiman, D. C. (2013). "MSiReader: An Open-Source Interface to View and Analyze High Resolving Power MS Imaging Files on Matlab Platform". Journal of the American Society for Mass Spectrometry. 24 (5): 718–721. PMC 3693088
 . PMID 23536269. doi:10.1007/s13361-013-0607-z.
. PMID 23536269. doi:10.1007/s13361-013-0607-z. - ↑ Race, A. M.; Palmer, A. D.; Dexter, A.; Steven, R. T.; Styles, I. B.; Bunch, J. (2016). "SpectralAnalysis: software for the masses". Analytical Chemistry. 88 (19): 9451–9458. PMID 27558772. doi:10.1021/acs.analchem.6b01643.
- ↑ Gamboa-Becerra, Roberto; Ramírez-Chávez, Enrique; Molina-Torres, Jorge; Winkler, Robert (2015-07-01). "MSI.R scripts reveal volatile and semi-volatile features in low-temperature plasma mass spectrometry imaging (LTP-MSI) of chilli (Capsicum annuum)". Analytical and Bioanalytical Chemistry. 407 (19): 5673–5684. PMID 26007697. doi:10.1007/s00216-015-8744-9.
- ↑ Bemis, Kyle D.; Harry, April; Eberlin, Livia S.; Ferreira, Christina; van de Ven, Stephanie M.; Mallick, Parag; Stolowitz, Mark; Vitek, Olga (2015-03-15). "Cardinal: an R package for statistical analysis of mass spectrometry-based imaging experiments". Bioinformatics. 31 (14): 2418–2420. PMC 4495298
 . PMID 25777525. doi:10.1093/bioinformatics/btv146.
. PMID 25777525. doi:10.1093/bioinformatics/btv146.