Holliday junction
.svg.png)
A Holliday junction is a branched nucleic acid structure that contains four double-stranded arms joined together. These arms may adopt one of several conformations depending on buffer salt concentrations and the sequence of nucleobases closest to the junction. The structure is named after the molecular biologist Robin Holliday, who proposed its existence in 1964.
In biology, Holliday junctions are a key intermediate in many types of genetic recombination, as well as in double-strand break repair. These junctions usually have a symmetrical sequence and are thus mobile, meaning that the four individual arms may slide through the junction in a specific pattern that largely preserves base pairing. Additionally, four-arm junctions similar to Holliday junctions appear in some functional RNA molecules.
Immobile Holliday junctions, with asymmetrical sequences that lock the strands in a specific position, were artificially created by scientists to study their structure as a model for natural Holliday junctions. These junctions also later found use as basic structural building blocks in DNA nanotechnology, where multiple Holliday junctions can be combined into specific designed geometries that provide molecules with a high degree of structural rigidity.
Structure

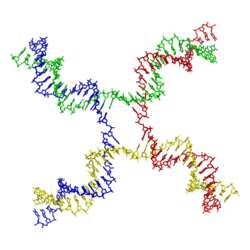

Holliday junctions may exist in a variety of conformational isomers with different patterns of coaxial stacking between the four double-helical arms. Coaxial stacking is the tendency of nucleic acid blunt ends to bind to each other, by interactions between the exposed bases. There are three possible stacking conformers: an unstacked form and two stacked forms. The unstacked form dominates in the absence of divalent cations such as Mg2+, because of electrostatic repulsion between the negatively charged backbones of the strands. In the presence of at least about 0.1 mM Mg2+, the electrostatic repulsion is counteracted and the stacked structures predominate. As of 2000, it was not known with certainty whether the electrostatic shielding was the result of site-specific binding of cations to the junction, or the presence of a diffuse collection of the ions in solution.[1]
The unstacked form is a nearly square planar, extended conformation. On the other hand, the stacked conformers have two continuous double-helical domains separated by an angle of about 60° in a right-handed direction. Two of the four strands stay roughly helical, remaining within each of the two double-helical domains, while the other two cross between the two domains in an antiparallel fashion.[1]
The two possible stacked forms differ in which pairs of the arms are stacked with each other; which of the two dominates is highly dependent on the base sequences nearest to the junction. Some sequences result in an equilibrium between the two conformers, while others strongly prefer a single conformer. In particular, junctions containing the sequence A-CC bridging the junction point appear to strongly prefer the conformer that allows a hydrogen bond to form between the second cytosine and one of the phosphates at the junction point. While most studies have focused on the identities of the four bases nearest to the junction on each arm, it is evident that bases farther out can also affect the observed stacking conformations.[1]
In junctions with symmetrical sequences, the branchpoint is mobile and can migrate in a random walk process. The rate of branch migration varies dramatically with ion concentration, with single-step times increasing from 0.3−0.4 ms with no ions to 270−300 ms with 10 mM Mg2+. The change in rate is correlated with the formation of the stacked versus the unstacked structures.[1]
Holliday junctions with a nick, or break in one of the strands, at the junction point adopt a perpendicular orientation, and always prefer the stacking conformer that places the nick on a crossover strand rather than a helical strand.[1]
RNA Holliday junctions assume an antiparallel stacked conformation at high magnesium concentrations, a perpendicular stacked conformation at moderate concentrations, and rotate into a parallel stacked conformation at low concentrations, while even small calcium ion concentrations favor the antiparallel conformer.[1]
Biological function
The Holliday junction is a key intermediate in homologous recombination, a biological process that increases genetic diversity by shifting genes between two chromosomes, as well as site-specific recombination events involving integrases. They are additionally involved in repair of double-strand breaks.[1] In addition, cruciform structures involving Holliday junctions can arise to relieve helical strain in symmetrical sequences in DNA supercoils.[2] While four-arm junctions also appear in functional RNA molecules, such as U1 spliceosomal RNA and the hairpin ribozyme of the tobacco ringspot virus, these usually contain unpaired nucleotides in between the paired double-helical domains, and thus do not strictly adopt the Holliday structure.[1]
The Holliday junctions in homologous recombination are between identical or nearly identical sequences, leading to a symmetric arrangement of sequences around the central junction. This allows a branch migration process to occur where the strands move through the junction point.[1] Cleavage, or resolution, of the Holliday junction can occur in two ways. Cleavage of the original set of strands leads to two molecules that may show gene conversion but not chromosomal crossover, while cleavage of the other set of two strands causes the resulting recombinant molecules to show crossover. All products, regardless of cleavage, are heteroduplexes in the region of Holliday junction migration.[3]
Many proteins are able to recognize or distort the Holliday junction structure. One such class contains junction-resolving enzymes that cleave the junctions, sometimes in a sequence-specific fashion. Such proteins distort the structure of the junction in various ways, often pulling the junction into an unstacked conformation, breaking the central base pairs, and/or changing the angles between the four arms. Other classes are branch migration proteins that increase the exchange rate by orders of magnitude, and site-specific recombinases.[1] In prokaryotes, Holliday junction resolvases fall into two families, integrases and nucleases, that are each structurally similar although their sequences are not conserved.[3]
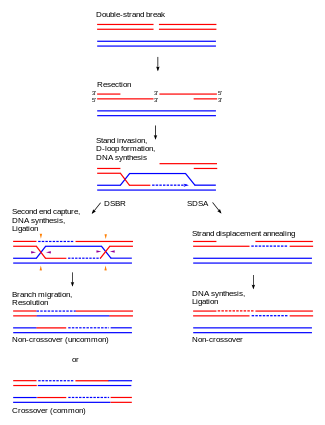
In eukaryotes, two primary models for how homologous recombination repairs double-strand breaks in DNA are the double-strand break repair (DSBR) pathway (sometimes called the double Holliday junction model) and the synthesis-dependent strand annealing (SDSA) pathway.[4] In the case of double strand breakage, the 3' end is degraded and the longer 5' end invades the contiguous sister chromatid, forming a replication bubble. As this bubble nears the broken DNA, the longer 5' antisense strand again invades the sense strand of this portion of DNA, transcribing a second copy. When replication ends, both tails are reconnected to form two Holliday Junctions, which are then cleaved in a variety of patterns by proteins.[5] An animation of this process can be seen here.[6]
Double-strand DNA breaks in bacteria are repaired by the RecBCD pathway of homologous recombination. Breaks that occur on only one of the two DNA strands, known as single-strand gaps, are thought to be repaired by the RecF pathway. Both the RecBCD and RecF pathways include a series of reactions known as branch migration, in which single DNA strands are exchanged between two intercrossed molecules of duplex DNA, and resolution, in which those two intercrossed molecules of DNA are cut apart and restored to their normal double-stranded state.[7] Homologous recombination occurs in several groups of viruses. In DNA viruses such as herpesvirus, recombination occurs through a break-and-rejoin mechanism like in bacteria and eukaryotes.[8] In bacteria, branch migration is facilitated by the RuvABC complex or RecG protein, molecular motors that use the energy of ATP hydrolysis to move the junction. The junction must then be resolved into two separate duplexes, restoring either the parental configuration or a crossed-over configuration. Resolution can occur in either a horizontal or vertical fashion during homologous recombination, giving patch products (if in same orientation during double strand break repair) or splice products (if in different orientations during double strand break repair).[9][10] RuvA and RuvB are branch migration proteins, while RuvC is a junction-resolving enzyme.[1]
There is evidence for recombination in some RNA viruses, specifically positive-sense ssRNA viruses like retroviruses, picornaviruses, and coronaviruses. There is controversy over whether homologous recombination occurs in negative-sense ssRNA viruses like influenza.[11]
Resolution
In budding yeast Saccharomyces cerevisiae, Holliday junctions can be resolved by four different pathways that account for essentially all Holliday junction resolution in vivo.[12] The pathway that produces the majority of crossovers in S. cerevisiae budding yeast, and possibly in mammals, involves proteins EXO1, MLH1-MLH3 heterodimer (called MutL gamma) and SGS1 (ortholog of Bloom syndrome helicase).[12] The MLH1-MLH3 heterodimer binds preferentially to Holliday junctions.[13] It is an endonuclease that makes single-strand breaks in supercoiled double-stranded DNA.[13][14] The MLH1-MLH3 heterodimer promotes the formation of crossover recombinants.[15] While the other three pathways, involving proteins MUS81-MMS4, SLX1 and YEN1, respectively, can promote Holliday junction resolution in vivo, absence of all three nucleases has only a modest impact on formation of crossover products.
Double mutants deleted for both MLH3 (major pathway) and MMS4 (minor pathway) showed dramatically reduced crossing over compared to wild-type (6- to 17-fold); however spore viability was reasonably high (62%) and chromosomal disjunction appeared mostly functional.[15]
Although MUS81 is a component of a minor crossover pathway in the meiosis of budding yeast, plants and vertebrates,[16] in the protozoan Tetrahymena thermophila, MUS81 appears to be part of an essential, if not the predominant crossover pathway.[16] The MUS81 pathway also appears to be the predominant crossover pathway in the fission yeast Schizosaccharomyces pombe.[16]
The MSH4 and MSH5 proteins form a hetero-oligomeric structure (heterodimer) in yeast and humans.[17][18][19] In the yeast Saccharomyces cerevisiae MSH4 and MSH5 act specifically to facilitate crossovers between homologous chromosomes during meiosis.[17] The MSH4/MSH5 complex binds and stabilizes double Holliday junctions and promotes their resolution into crossover products. An MSH4 hypomorphic (partially functional) mutant of S. cerevisiae showed a 30% genome wide reduction in crossover numbers, and a large number of meioses with non exchange chromosomes.[20] Nevertheless, this mutant gave rise to spore viability patterns suggesting that segregation of non-exchange chromosomes occurred efficiently. Thus in S. cerevisiae proper segregation apparently does not entirely depend on crossovers between homologous pairs.
Use in DNA nanotechnology

DNA nanotechnology is the design and manufacture of artificial nucleic acid structures as engineering materials for nanotechnology rather than as the carriers of genetic information in living cells. The field uses branched DNA structures as fundamental components to create more complex, rationally designed structures. Holliday junctions are thus components of many such DNA structures. As isolated Holliday junction complexes are too flexible to assemble into large ordered arrays, structural motifs with multiple Holliday junctions are used to create rigid "tiles" that can then assemble into larger "arrays".[22][23]
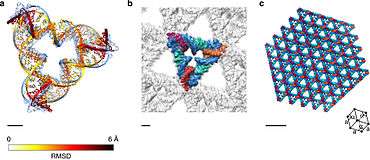
The most common such motif is the double crossover (DX) complex, which contains two Holliday junctions in close proximity to each other, resulting in a rigid structure that can self-assemble into larger arrays. The structure of the DX molecule forces the Holliday junctions to adopt a conformation with the double-helical domains directly side-by-side, in contrast to their preferred angle of about 60°. The complex can be designed to force the junctions into either a parallel or antiparallel orientation, but in practice the antiparallel variety are more well-behaved, and the parallel version is rarely used.[22][23]
The DX structural motif is the fundamental building block of the DNA origami method, which is used to make larger two- and three-dimensional structures of arbitrary shape. Instead of using individual DX tiles, a single long scaffold strand is folded into the desired shape by a number of short staple strands. When assembled, the scaffold strand is continuous through the double-helical domains, while the staple strands participate in the Holliday junctions as crossover strands.[25]
Some tile types that retain the Holliday junction's native 60° angle have been demonstrated. One such array uses tiles containing four Holliday junctions in a parallelogram arrangement. This structure had the benefit of allowing the junction angle to be directly visualized via atomic force microscopy. Tiles of three Holliday junctions in a triangular fashion have been used to make periodic three-dimensional arrays for use in X-ray crystallography of biomolecules. These structures are named for their similarity to structural units based on the principle of tensegrity, which utilizes members both in tension and compression.[22][23]
History
Robin Holliday proposed the junction structure that now bears his name as part of his model of homologous recombination in 1964, based on his research on the organisms Ustilago maydis and Saccharomyces cerevisiae. The model provided a molecular mechanism that explained both gene conversion and chromosomal crossover. Holliday realized that the proposed pathway would create heteroduplex DNA segments with base mismatches between different versions of a single gene. He predicted that the cell would have a mechanism for mismatch repair, which was later discovered.[3] Prior to Holliday's model, the accepted model involved a copy-choice mechanism[26] where the new strand is synthesized directly from parts of the different parent strands.[27]
In the original Holliday model for homologous recombination, single-strand breaks occur at the same point on one strand of each parental DNA. Free ends of each broken strand then migrate across to the other DNA helix. There, the invading strands are joined to the free ends they encounter, resulting in the Holliday junction. As each crossover strand reanneals to its original partner strand, it displaces the original complementary strand ahead of it. This causes the Holliday junction to migrate, creating the heteroduplex segments. Depending on which strand was used as a template to repair the other, the four cells resulting from meiosis might end up with three copies of one allele and only one of the other, instead of the normal two of each, a property known as gene conversion.[3]
Holliday's original model assumed that heteroduplex DNA would be present on both chromosomes, but experimental data on yeast refuted this. An updated model by Matt Meselson and Charley Radding in 1975 introduced the idea of branch migration.[26] Further observations in the 1980s led to the proposal of alternate mechanisms for recombination such as the double-strand break model (by Jack Szostak, Frank Stahl, and others) and the single-strand annealing model. A third, the synthesis-dependent strand annealing model, did not involve Holliday junctions.[3]
The first experimental evidence for the structure of the Holliday junction came from electron microscopy studies in the late 1970s, where the four-arm structure was clearly visible in images of plasmid and bacteriophage DNA. Later in the 1980s, enzymes responsible for initiating the formation of, and binding to, Holliday junctions were identified, although as of 2004 the identification of mammalian Holliday junction resolvases remained elusive (however, see section "Resolution of Holliday junctions," above for more recent information). In 1983, artificial Holliday junction molecules were first constructed from synthetic oligonucleotides by Nadrian Seeman, allowing for more direct study of their physical properties. Much of the early analysis of Holliday junction structure was inferred from gel electrophoresis, FRET, and hydroxyl radical and nuclease footprinting studies. In the 1990s, crystallography and nucleic acid NMR methods became available, as well as computational molecular modelling tools.[1][3][28]
Initially, geneticists assumed that the junction would adopt a parallel rather than antiparallel conformation, because that would place the homologous duplexes in closer alignment to each other.[1] Chemical analysis in the 1980s showed that the junction actually preferred the antiparallel conformation, a finding that was considered controversial, and Robin Holliday himself initially doubted the findings.[1][3] The antiparallel structure later became widely accepted due to X-ray crystallography data on in vitro molecules, although as of 2004 the implications for the in vivo structure remained unclear, especially the structure of the junctions is often altered by proteins bound to it.[3]
The conceptual foundation for DNA nanotechnology was first laid out by Nadrian Seeman in the early 1980s.[29] A number of natural branched DNA structures were known at the time, including the DNA replication fork and the mobile Holliday junction, but Seeman's insight was that immobile nucleic acid junctions could be created by properly designing the strand sequences to remove symmetry in the assembled molecule, and that these immobile junctions could in principle be combined into rigid crystalline lattices. The first theoretical paper proposing this scheme was published in 1982, and the first experimental demonstration of an immobile DNA junction was published the following year.[23][30] Seeman developed the more rigid double-crossover (DX) motif, suitable for forming two-dimensional lattices, demonstrated in 1998 by him and Erik Winfree.[22] In 2006, Paul Rothemund first demonstrated the DNA origami technique for easily and robustly creating folded DNA structures of arbitrary shape. This method allowed the creation of much larger structures than were previously possible, and which are less technically demanding to design and synthesize.[31] The synthesis of a three-dimensional lattice was finally published by Seeman in 2009, nearly thirty years after he had set out to achieve it.[32]
References
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 Lilley, David M. J. (2000). "Structures of helical junctions in nucleic acids". Quarterly reviews of biophysics. 33 (2): 109–159. PMID 11131562. doi:10.1017/S0033583500003590.
- ↑ Bloomfield, Victor A.; Crothers, Donald M.; Tinoco, Jr., Ignacio (2000). Nucleic acids: structures, properties, and functions. Sausalito, California: University Science Books. p. 468. ISBN 0935702490.
- 1 2 3 4 5 6 7 8 Liu Y, West S (2004). "Happy Hollidays: 40th anniversary of the Holliday junction". Nature Reviews Molecular Cell Biology. 5 (11): 937–44. PMID 15520813. doi:10.1038/nrm1502.
- ↑ Sung, P; Klein, H (October 2006). "Mechanism of homologous recombination: mediators and helicases take on regulatory functions". Nature Reviews Molecular Cell Biology. 7 (10): 739–750. PMID 16926856. doi:10.1038/nrm2008.
- ↑ Hartel, Daniel L.; Jones, Elizabeth W. (2009). "Chapter 6: Molecular Biology of DNA Replication and Recombination". Genetics: Analysis of Genetics and Genomes. Burlington: Jones & Bartlett.
- ↑ Helleday, T. "Double-Strand Break Repair via Double Holliday Junctions (Szostak Model)". Animation. MIT.
- ↑ Rocha, EPC; Cornet, E; Michel, B (August 2005). "Comparative and evolutionary analysis of the bacterial homologous recombination systems". PLoS Genetics. 1 (2): e15. PMC 1193525
 . PMID 16132081. doi:10.1371/journal.pgen.0010015.
. PMID 16132081. doi:10.1371/journal.pgen.0010015. 
- ↑ Fleischmann Jr, WR (1996). "Chapter 43". Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 0-9631172-1-1.
- ↑ West SC (2003). "Molecular views of recombination proteins and their control". Nature Reviews Molecular Cell Biology. 4 (6): 435–45. PMID 12778123. doi:10.1038/nrm1127.
- ↑ Kowalczykowski SC (2000). "Initiation of genetic recombination and recombination-dependent replication". Trends in Biochemical Sciences. 25 (4): 156–65. PMID 10754547. doi:10.1016/S0968-0004(00)01569-3.
- ↑ Boni, MF; de Jong, MD; van Doorn, HR; Holmes, EC; Martin, Darren P. (3 May 2010). Martin, Darren P., ed. "Guidelines for identifying homologous recombination events in influenza a virus". PLoS ONE. 5 (5): e10434. PMC 2862710
 . PMID 20454662. doi:10.1371/journal.pone.0010434.
. PMID 20454662. doi:10.1371/journal.pone.0010434. 
- 1 2 Zakharyevich, K; Tang, S; Ma, Y; Hunter, N (April 2012). "Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase". Cell. 149 (2): 334–47. PMC 3377385
 . PMID 22500800. doi:10.1016/j.cell.2012.03.023.
. PMID 22500800. doi:10.1016/j.cell.2012.03.023. - 1 2 Ranjha L, Anand R, Cejka P. 2014. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to Holliday junctions. J Biol Chem. 2014 Feb 28;289(9):5674-86. doi:10.1074/jbc.M113.533810. PMID 24443562
- ↑ Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E (2014). "Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease". J. Biol. Chem. 289 (9): 5664–73. PMC 3937641
 . PMID 24403070. doi:10.1074/jbc.M113.534644.
. PMID 24403070. doi:10.1074/jbc.M113.534644. - 1 2 Sonntag Brown M, Lim E, Chen C, Nishant KT, Alani E (2013). "Genetic analysis of mlh3 mutations reveals interactions between crossover promoting factors during meiosis in baker's yeast". G3: Genes, Genomes, Genetics. 3 (1): 9–22. PMC 3538346
 . PMID 23316435. doi:10.1534/g3.112.004622.
. PMID 23316435. doi:10.1534/g3.112.004622. - 1 2 3 Lukaszewicz A, Howard-Till RA, Loidl J (2013). "Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex". Nucleic Acids Res. 41 (20): 9296–309. PMC 3814389
 . PMID 23935123. doi:10.1093/nar/gkt703.
. PMID 23935123. doi:10.1093/nar/gkt703. - 1 2 Pochart P, Woltering D, Hollingsworth NM (1997). "Conserved properties between functionally distinct MutS homologs in yeast". J. Biol. Chem. 272 (48): 30345–9. PMID 9374523. doi:10.1074/jbc.272.48.30345.
- ↑ Winand NJ, Panzer JA, Kolodner RD (1998). "Cloning and characterization of the human and Caenorhabditis elegans homologs of the Saccharomyces cerevisiae MSH5 gene". Genomics. 53 (1): 69–80. PMID 9787078. doi:10.1006/geno.1998.5447.
- ↑ Bocker T, Barusevicius A, Snowden T, Rasio D, Guerrette S, Robbins D, et al. (1999). "hMSH5: a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis". Cancer Res. 59 (4): 816–22. PMID 10029069.
- ↑ Krishnaprasad GN, Anand MT, Lin G, Tekkedil MM, Steinmetz LM, Nishant KT (2015). "Variation in crossover frequencies perturb crossover assurance without affecting meiotic chromosome segregation in Saccharomyces cerevisiae". Genetics. 199 (2): 399–412. PMC 4317650
 . PMID 25467183. doi:10.1534/genetics.114.172320.
. PMID 25467183. doi:10.1534/genetics.114.172320. - ↑ Mao, Chengde (December 2004). "The emergence of complexity: lessons from DNA". PLoS Biology. 2 (12): 2036–2038. PMC 535573
 . PMID 15597116. doi:10.1371/journal.pbio.0020431.
. PMID 15597116. doi:10.1371/journal.pbio.0020431. - 1 2 3 4 Seeman, Nadrian C. (June 2004). "Nanotechnology and the double helix". Scientific American. 290 (6): 64–75. PMID 15195395. doi:10.1038/scientificamerican0604-64.
- 1 2 3 4 Seeman, Nadrian C. (2010). "Nanomaterials based on DNA". Annual Review of Biochemistry. 79: 65–87. PMC 3454582
 . PMID 20222824. doi:10.1146/annurev-biochem-060308-102244.
. PMID 20222824. doi:10.1146/annurev-biochem-060308-102244. - ↑ Pan, Keyao; Kim, Do-Nyun; Zhang, Fei; Adendorff, Matthew R.; Yan, Hao; Bathe, Mark (2014-12-03). "Lattice-free prediction of three-dimensional structure of programmed DNA assemblies". Nature Communications. 5: 5578. PMC 4268701
 . PMID 25470497. doi:10.1038/ncomms6578.
. PMID 25470497. doi:10.1038/ncomms6578. - ↑ Saccà, Barbara; Niemeyer, Christof M. (2012). "DNA Origami: The Art of Folding DNA" (PDF). Angewandte Chemie International Edition. 51 (1): 58–66. doi:10.1002/anie.201105846. Retrieved 25 February 2015.
- 1 2 Stahl FW (1 October 1994). "The Holliday junction on its thirtieth anniversary" (PDF). Genetics. 138 (2): 241–246. PMC 1206142
 . PMID 7828807.
. PMID 7828807. - ↑ Advances in genetics. Academic Press. 1971. ISBN 9780080568027.
- ↑ Hays FA, Watson J, Ho PS (2003). "Caution! DNA Crossing: Crystal Structures of Holliday Junctions". J Biol Chem. 278 (50): 49663–49666. PMID 14563836. doi:10.1074/jbc.R300033200.
- ↑ Pelesko, John A. (2007). Self-assembly: the science of things that put themselves together. New York: Chapman & Hall/CRC. pp. 201, 242, 259. ISBN 978-1-58488-687-7.
- ↑ Pinheiro, A. V.; Han, D.; Shih, W. M.; Yan, H. (December 2011). "Challenges and opportunities for structural DNA nanotechnology". Nature Nanotechnology. 6 (12): 763–772. PMC 3334823
 . PMID 22056726. doi:10.1038/nnano.2011.187.
. PMID 22056726. doi:10.1038/nnano.2011.187. - ↑ Rothemund, Paul W. K. (2006). "Scaffolded DNA origami: from generalized multicrossovers to polygonal networks". In Chen, Junghuei; Jonoska, Natasha; Rozenberg, Grzegorz. Nanotechnology: science and computation. Natural Computing Series. New York: Springer. pp. 3–21. ISBN 978-3-540-30295-7. doi:10.1007/3-540-30296-4_1.
- ↑ Service, Robert F. (3 June 2011). "DNA nanotechnology grows up". Science. 332 (6034): 1140–1143. PMID 21636754. doi:10.1126/science.332.6034.1140.
External links
- Holliday junctions at the US National Library of Medicine Medical Subject Headings (MeSH)
- Conformational Change of Holliday Junction
- Analysis of branch migration activities of proteins using synthetic DNA substrates (a protocol)