HMG-box
| HMG (high mobility group) box | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
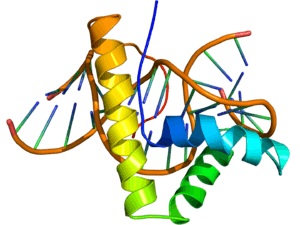
NMR structure of the HMG-box domain of the LEF1 protein (rainbow colored, N-terminus = blue, C-terminus = red) complexed with DNA (brown) based on the PDB: 2LEF coordinates. | |||||||||
| Identifiers | |||||||||
| Symbol | PF00505 | ||||||||
| Pfam | PF00505 | ||||||||
| InterPro | IPR009071 | ||||||||
| SCOP | 1hsm | ||||||||
| SUPERFAMILY | 1hsm | ||||||||
| |||||||||
In molecular biology, the HMG-box (high mobility group box) is a protein domain which is involved in DNA binding.[1]
Structure
The structure of the HMG-box domain contains three alpha helices separated by loops (see figure to the right).[2]
Function
HMG-box containing proteins only bind non-B-type DNA conformations (kinked or unwound) with high affinity.[1] HMG-box domains are found in high mobility group proteins, which are involved in the regulation of DNA-dependent processes such as transcription, replication, and DNA repair, all of which require changing the conformation of chromatin.[2] The single and the double box HMG proteins alter DNA architecture by inducing bends upon binding.[3][4]
References
- 1 2 Stros M, Launholt D, Grasser KD (October 2007). "The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins". Cell. Mol. Life Sci. 64 (19–20): 2590–606. PMID 17599239. doi:10.1007/s00018-007-7162-3.
- 1 2 Thomas JO (August 2001). "HMG1 and 2: architectural DNA-binding proteins". Biochem. Soc. Trans. 29 (Pt 4): 395–401. PMID 11497996. doi:10.1042/BST0290395.
- ↑ D. Murugesapillai et al, DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin, Nucl Acids Res (2014) 42 (14): 8996-9004
- ↑ D. Murugesapillai et al, Single-molecule studies of high-mobility group B architectural DNA bending proteins, Biophys Rev (2016) doi:10.1007/s12551-016-0236-4
External links
- HMG-Box Domains at the US National Library of Medicine Medical Subject Headings (MeSH)
This article is issued from
Wikipedia.
The text is licensed under Creative Commons - Attribution - Sharealike.
Additional terms may apply for the media files.