Genetics and archaeogenetics of South Asia
The study of the genetics and archaeogenetics of the ethnic groups of South Asia aims at uncovering these groups' genetic history. The geographic position of the Indian subcontinent makes its biodiversity important for the study of the early dispersal of anatomically modern humans across Asia.
Studies based on mtDNA variation have reported genetic unity across various Indian sub–populations.[1][2][3][4] Conclusions of studies based on Y Chromosome variation and Autosomal DNA variation have been varied, although many researchers argue that most of the ancestral nodes of the phylogenetic tree of all the mtDNA types originated in the subcontinent. Recent genome studies appear to show that most South Asians are descendants of two major ancestral components, one restricted to South Asia (Ancestral South Indian) and the other component (Ancestral North Indian) more closely related to those in Central Asia, West Asia and Europe.[5][6]
It has been found that the ancestral node of the phylogenetic tree of all the mtDNA types typically found in Central Asia, the West Asia and Europe are also to be found in South Asia at relatively high frequencies. The inferred divergence of this common ancestral node is estimated to have occurred slightly less than 50,000 years ago.[7] In India the major maternal lineages, or mitochondrial DNA haplogroups are M, R and U, whose coalescence times have been approximated to 50,000 BP.[7]
The major paternal lineages represented by Y chromosomes are haplogroups R1a1, R2, H, L and J2.[8] Many researchers have argued that Y-DNA Haplogroup R1a1 (M17) is of autochthonous Indian origin.[9][10] However, proposals for a Central Asian origin for R1a1 are also quite common.[11][12]
Overview
All the mtDNA and Y-chromosome lineages outside Africa descend from three founder lineages:
All these six founder haplogroups can be found in the present day populations of South Asia. Moreover, the mtDNA haplogroup M and the Y-chromosome haplogroups C and D are restricted to the area east of South Asia. All the West Eurasian populations derive from the N and R haplogroups of mtDNA and the F haplogroup of the Y-chromosome.[13]
Endicott et al. state that these facts are consistent with the hypothesis of a single exodus from East Africa 65,000 years ago via a southern coastal route, with the West Eurasian lineages separating from the South Asian lineages somewhere between East/Northeast Africa and South Asia.[14]
mtDNA
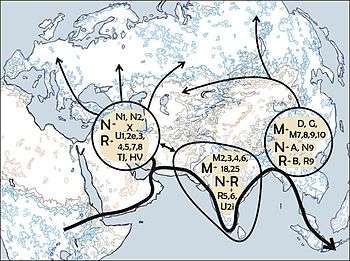
The most frequent mtDNA haplogroups in the Indian subcontinent are M, R and U (where U is a descendant of R).[8]
Arguing for the longer term "rival Y-Chromosome model",[9] Stephen Oppenheimer believes that it is highly suggestive that India is the origin of the Eurasian mtDNA haplogroups which he calls the "Eurasian Eves". According to Oppenheimer it is highly probable that nearly all human maternal lineages in Central Asia, the Middle East and Europe descended from only four mtDNA lines that originated in South Asia 50,000-100,000 years ago.[15]
Macrohaplogroup M
The macrohaplogroup M which is considered as a cluster of the proto-Asian maternal lineages,[7] represents more than 60% of Indian MtDNA.[16]
The M macrohaplotype in India includes many subgroups that differ profoundly from other sublineages in East Asia especially Mongoloid populations.[7] The deep roots of M phylogeny clearly ascertain the relic of Indian lineages as compared to other M sub lineages (in East Asia and elsewhere) suggesting 'in-situ' origin of these sub-haplogroups in South Asia, most likely in India. These deep rooting lineages are not language specific and spread over all the language groups in India.[16]
Virtually all modern Central Asian MtDNA M lineages seem to belong to the Eastern Eurasian (Mongolian) rather than the Indian subtypes of haplogroup M, which indicates that no large-scale migration from the present Turkic-speaking populations of Central Asia occurred to India. The absence of haplogroup M in Europeans, compared to its equally high frequency among Indians, eastern Asians and in some Central Asian populations contrasts with the Western Eurasian leanings of South Asian paternal lineages.[7]
Most of the extant mtDNA boundaries in South and Southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans.[17]
| Haplogroup | Important Sub clades | Populations |
|---|---|---|
| M2 | M2a, M2b | Throughout the continent with low presence in Northwest Peaking in Bangladesh, Andhra Pradesh, coastal Tamil Nadu and Sri Lanka |
| M3 | M3a | Concentrated into northwestern India Highest amongst the Parsees of Mumbai |
| M4 | M4a | Peaks in Pakistan, Kashmir and Andhra Pradesh |
| M6 | M6a, M6b | Kashmir and near the coasts of the Bay of Bengal, Sri Lanka |
| M18 | Throughout the subcontinent Peaking at Rajasthan and Andhra Pradesh | |
| M25 | Moderately frequent in Kerala and Maharashtra but rather infrequent elsewhere in India |
Macrohaplogroup R
The macrohaplogroup R (a very large and old subdivision of macrohaplogroup N) is also widely represented and accounts for the other 40% of Indian MtDNA. A very old and most important subdivision of it is haplogroup U that, while also present in West Eurasia, has several subclades specific to South Asia.
Most important South Asian haplogroups within R:[17]
| Haplogroup | Populations |
|---|---|
| R2 | Distributed widely across the sub continent |
| R5 | widely distributed by most of India. Peaks in coastal SW India |
| R6 | widespread at low rates across India. Peaks among Tamils and Kashmiris |
| W | Found in northwestern states. Peaks in Gujarat, Punjab and Kashmir, frequency is low elsewhere. |
Haplogroup U
Haplogroup U is a sub-haplogroup of macrohaplogroup R.[17] The distribution of haplogroup U is a mirror image of that for haplogroup M: the former has not been described so far among eastern Asians but is frequent in European populations as well as among Indians.[18] Indian U lineages differ substantially from those in Europe and their coalescence to a common ancestor also dates back to about 50,000 years.[1]
| Haplogroup | Populations |
|---|---|
| U2* | (a parahaplogroup) is sparsely distributed specially in the northern half of the subcontinent.
It is also found in SW Arabia. |
| U2a | shows relatively high density in Pakistan and NW India but also in Karnataka, where it reaches its higher density. |
| U2b | has highest concentration in Uttar Pradesh but is also found in many other places, specially in Kerala and Sri Lanka.
It is also found in Oman. |
| U2c | is specially important in Bangladesh and West Bengal. |
| U2l | is maybe the most important numerically among U subclades in South Asia, reaching specially high concentrations (over 10%) in Uttar Pradesh, Sri Lanka, Sindh and parts of Karnataka. It also has some importance in Oman. mtDNA haplogroup U2i is dubbed "Western Eurasian" in Bamshad et al. study but "Eastern Eurasian (mostly India specific)" in Kivisild et al. study. |
| U7 | this haplogroup has a significant presence in Gujarat, Punjab and Pakistan. The possible homeland of this haplogroup spans Indian Gujarat (highest frequency, 12%) and Iran because from there its frequency declines steeply both to the east and to the west. |
Y chromosome
The major Y chromosome DNA haplogroups in the subcontinent are Haplogroup F's descendant haplogroups R (mostly R1a1, R2 and R2a), L, H and J (mostly J2).[8]
The South Asian Y-chromosomal gene pool is characterized by five major lineages: R1a, R2, H, L and J2. Their geographical origins are listed as follows, according to the latest scholarship:
| Major South Asian Y-chromosomal lineages: | H | J2 | L | R1a | R2 |
|---|---|---|---|---|---|
| Basu et al. (2003) | no comment | no comment | no comment | Central Asia | no comment |
| Kivisild et al. (2003) | India | Western Asia | India | Southern and Western Asia | South-Central Asia |
| Cordaux et al. (2004) | India | West or Central Asia | Middle Eastern | Central Asia | South-Central Asia |
| Sengupta et al. (2006) | India | The Middle East and Central Asia | South India | North India | North India |
| Thanseem et al. (2006) | India | The Levant | The Middle East | Southern and Central Asia | Southern and Central Asia |
| Sahoo et al. (2006) | South Asia | The Near East | South Asia | South or West Asia | South Asia |
| Mirabal et al. (2009) | no comment | no comment | no comment | Northwestern India or Central Asia | no comment |
| Zhao et al. (2009) | India | The Middle East | The Middle East | Central Asia or West Eurasia | Central Asia or West Eurasia |
| Sharma et al. (2009) | no comment | no comment | no comment | South Asia | no comment |
| Thangaraj et al. (2010) | South Asia | The Near East | The Near East | South Asia | South Asia |
Haplogroup H
Haplogroup H (Y-DNA) is found at a high frequency in South Asia. H is rarely found outside of the South Asia but is common among the Romanis, particularly the H-M82 subgroup. Haplogroup H is frequently found among populations of India, Sri Lanka, Nepal, Pakistan and Maldives. All three branches of Haplogroup H (Y-DNA) are found in Indian-subcontinent.
It is a branch of Haplogroup F and descends from GHIJK family. Haplogroup H is believed to have arisen in South Asia between 30,000 and 40,000 years ago.[19] Its probable site of introduction is South Asia, since it is concentrated there. It seems to represent the main Y-Chromosome haplogroup of the paleolithic inhabitants of Indian-Subcontinent. Some individuals in South Asia have also been shown to belong to the much rarer subclade H3 (Z5857).[19] Haplogroup H is by no means restricted to specific populations. For example, H is possessed by about 28.8% of Indo-Aryan castes.[9][20] and in tribals about 25-35%.[11][20]
Haplogroup J2
Haplogroup J2 reflects presence from neolithic period in the subcontinent.[21] The frequency of J2 is higher in South Indian castes (19%) than in North Indian castes (11%) or Pakistan (12%).[9] Haplogroup J2 frequency is higher among south Indian middle castes at 21%, followed by upper castes at 18.6%, and lower castes 14%.[9] J2 is present in tribals too and has a frequency of 11% in Austro-Asiatic tribals. Among the Austro-Asiatic tribals, the predominant J2 occurs in the Lodha(35%).[9] J2 is also present in the South Indian hill tribe Toda at a frequency of 38.46%[22] and in the Kol tribe of Uttar Pradesh at a frequency of 33.34%.[23] Haplogroup J-P209 was found to be more common in India's Shia Muslims, of which 28.7% belong to haplogroup J, with 13.7% in J-M410, 10.6% in J-M267 and 4.4% in J2b (Eaaswarkhanth 2009).
In Pakistan, the highest frequencies of J2-M172 were observed among the Parsis at 38.89%, the Dravidian speaking Brahui's at 28.18% and the Makrani Balochs at 24%.[24] It also occurs at 18.18% in Makrani Siddis and at 3% in Karnataka Siddis.[24][25]
J2-M172 is found at an overall frequency of 16.1% in the people of Sri Lanka.[26] In Maldives, 22% of Maldivian population were found to be haplogroup J2 positive.
Haplogroup L
India
Haplogroup L shows time of neolithic expansion.[21] The clade is present in the Indian population at an overall frequency of ca.7-15%.[9][11][27][20] There are three subbranches of Haplogroup L and all three are found mostly in South Asia. Haplogroup L has higher frequency among south Indian castes (ca. 17-19%) and reaches up to 68% in some castes in Karnataka but is somewhat rarer in north Indian castes (ca. 5-6%).[9][28] They make a case for an indigenous origin of L-M76 in South Asia as the spatial distributions of both L-M76 HG frequency and associated microsatellite variance show a pattern of spread emanating from southern India.[9] The presence of haplogroup L is quite rare among tribal groups (ca. 5,6-7%)[9][11][20]
Pakistan
Haplogroup L3 (M357) is found frequently among Burusho (approx. 12%[29]) and Pashtuns (approx. 7%[29]). Its highest frequency can be found in south western Balochistan province along the Makran coast (28%) to Indus River delta. L3a (PK3) is found in approximately 23% of Nuristani in northwest Pakistan.[29]
The clade is present in moderate distribution among the general Pakistani population (approx. 11.6%[29]).
Haplogroup R1a1
In South Asia R1a1 has been observed often with high frequency in a number of demographic groups,[10][30][31] as well as with highest STR diversity which lead some to see it as the locus of origin.[32][33][34]
While R1a originated ca. 22,000[33] to 25,000[35] years ago, its subclade M417 (R1a1a1) diversified ca. 5,800 years ago.[35] The distribution of M417-subclades R1-Z282 (including R1-Z280)[36] in Central- and Eastern Europe and R1-Z93 in Asia[36][35] suggests that R1a1a diversified within the Eurasian Steppes or the Middle East and Caucasus region.[36] The place of origin of these subclades plays a role in the debate about the origins of Indo-Europeans.
India
In India, high percentage of this haplogroup is observed in West Bengal Brahmins (72%) [30] to the east, Konkanastha Brahmins (48%) [30] to the west, Khatris (67%)[31] in north, Iyengar Brahmins (31%) in the south.[30] It has also been found in several South Indian Dravidian-speaking Tribals including the Chenchu (26%)[32] and Valmikis of Andhra Pradesh as well as the Yadav and Kallar of Tamil Nadu suggesting that M17 is widespread in these Southern Indians tribes.[32] Besides these, studies show high percentages in regionally diverse groups such as Manipuris (50%) [31] to the extreme North East and in among Punjabis (47%)[32] to the extreme North West.
Pakistan
In Pakistan it is found at 71% among the Mohanna of Sindh Province to the south and 46% among the Baltis of Gilgit-Baltistan to the north.[31]
Sri Lanka
23% of the Sinhalese people out of a sample of 87 subjects were found to be R1a1a (R-SRY1532) positive according to a 2003 research.[32]
Maldives
In Maldives, 24% of the Maldivian people were found to be R1a1a (M17) positive.
Nepal
People in Terai Region, Nepal show R1a1a at 69%.[37]
Haplogroup R2
In South Asia, the frequency of R2 and R2a lineage is around 10-15% in India and Sri Lanka and 7-8% in Pakistan. At least 90% of R-M124 individuals are located in the Indian sub-continent.[38] It is also reported in Caucasus and Central Asia at lower frequency.
India
Among regional groups, it is found among West Bengalis (23%), New Delhi Hindus (20%), Punjabis (5%) and Gujaratis (3%).[32] Among tribal groups, Karmalis of West Bengal showed highest at 100%[10] followed by Lodhas (43%)[39] to the east, while Bhil of Gujarat in the west were at 18%,[33] Tharus of north showed it at 17%,[4] Chenchu and Pallan of south were at 20% and 14% respectively.[10][9] Among caste groups, high percentages are shown by Jaunpur Kshatriyas (87%), Kamma Chaudhary (73%), Bihar Yadav (50%), Khandayat (46%)and Kallar (44%).[10]
It is also significantly high in many Brahmin groups including Punjabi Brahmins (25%), Bengali Brahmins (22%), Konkanastha Brahmins (20%), Chaturvedis (32%), Bhargavas (32%), Kashmiri Pandits (14%) and Lingayat Brahmins (30%).[33][12][4][10]
North Indian Muslims have a frequency of 19% (Sunni) and 13% (Shia),[12] while Dawoodi Bohra Muslim in the western state of Gujarat have a frequency of 16% and Mappla Muslims of South India have a frequency of 5%.[40]
Pakistan
The R2 haplogroup is found in 14% of the Burusho people.[29] Among the Hunza it is found at 18% while the Parsis show it at 20%. It is also found in the northeastern part of Afghanistan.
Sri Lanka
39% of the Sinhalese of Sri Lanka were found to be R2 positive according to a 2003 research.[32]
Maldives
13% of the Maldivian people of Maldives are found to have R2
Nepal
In Nepal, R2 percentages range from 2% to 26% within different groups under various studies. Newars show a significantly high frequency of 26% while people of Kathmandu show it at 10%.
Reconstructing Indian population history
The Indian Genome Variation Consortium (2008), divides the population of the subcontinent into four linguistic groups— Indo-European, Dravidian, Tibeto-Burman and Austro-Asiatic.[41][42][43][44][45] The molecular anthropology studies use three different type of markers: Mitochondrial DNA (mtDNA) variation which is maternally inherited and highly polymorphic, Y Chromosome variation which involves uniparental transmission along the male lines, and Autosomal DNA variation.[4]:04
mtDNA variation
Most of the studies based on mtDNA variation have reported genetic unity of Indian populations across language, caste and tribal groups.[1][2][3] It is likely that haplogroup M was brought to Asia from East Africa along the southern route by earliest migration wave 60,000 years ago.[1]
According to Kivisild et al. (1999), "Minor overlaps with lineages described in other Eurasian populations clearly demonstrate that recent immigrations have had very little impact on the innate structure of the maternal gene pool of Indians. Despite the variations found within India, these populations stem from a limited number of founder lineages. These lineages were most likely introduced to the Indian subcontinent during the Middle Palaeolithic, before the peopling of Europe and perhaps the Old World in general."[1] Basu et al. (2003) also emphasizes underlying unity of female lineages in India.[27]
Y Chromosome variation
Conclusions based on Y Chromosome variation have been more varied than those based on mtDNA variation. While Kivisild et al. (2003) proposes an ancient and shared genetic heritage of male lineages in India, Bamshad et al. (2001) suggests an affinity between Indian male lineages and west Eurasians proportionate to caste rank and places caste populations of southern Indian states closer to East Europeans.[46]
Basu et al. (2003) concludes that Austro–Asiatic tribal populations entered India first from the Northwest corridor and much later some of them through Northeastern corridor.[27] Whereas, Kumar et al. (2007) analyzed 25 Indian Austro-Asiatic tribes and found strong paternal genetic link among the sub-linguistic groups of the Indian Austro-Asiatic populations.[39] Mukherjee et al. (2001) places North Indians between west Asian and Central Asian populations,[47] whereas Cordaux et al. (2004) argues that the Indian caste populations are closer to Central Asian populations.[20] Sahoo et al. (2006) and Sengupata et al. (2006) suggest that Indian caste populations have not been subject to any recent admixtures.[9][10] Sanghamitra Sahoo concludes his study with:[10]
It is not necessary, based on the current evidence, to look beyond South Asia for the origins of the paternal heritage of the majority of Indians at the time of the onset of settled agriculture. The perennial concept of people, language, and agriculture arriving to India together through the northwest corridor does not hold up to close scrutiny. Recent claims for a linkage of haplogroups J2, L, R1a, and R2 with a contemporaneous origin for the majority of the Indian castes’ paternal lineages from outside the subcontinent are rejected, although our findings do support a local origin of haplogroups F* and H. Of the others, only J2 indicates an unambiguous recent external contribution, from West Asia rather than Central Asia. The current distributions of haplogroup frequencies are, with the exception of the lineages, predominantly driven by geographical, rather than cultural determinants. Ironically, it is in the northeast of India, among the TB groups that there is clear-cut evidence for large-scale demic diffusion traceable by genes, culture, and language, but apparently not by agriculture.
Autosomal DNA variation
Results of studies based upon autosomal DNA variation have also been varied. In a major study (2009) using over 500,000 biallelic autosomal markers, Reich hypothesized that the modern Indian population was the result of admixture between two genetically divergent ancestral populations dating from the post-Holocene era. These two "reconstructed" ancient populations he termed "Ancestral South Indians" (ASI) and "Ancestral North Indians" (ANI). According to Reich: "ANI ancestry is significantly higher in Indo-European than Dravidian speakers, suggesting that the ancestral ASI may have spoken a Dravidian language before mixing with the ANI."[48]
Further building on Reich et al.'s characterization of the South Asian population as historically based on admixture of ANI (Ancestral North Indian) and ASI (Ancestral South Indian) populations, a 2013 paper by Moorjani et al. states that a major mixture between populations in India occurred 1,900–4,200 years BP characterized by the deurbanization of the Indus civilization and population shift to the Gangetic system.[6]
Basu et al. (2003) suggests concludes that "Dravidian tribals were possibly widespread throughout India before the arrival of the Indo-European-speaking nomads" and that "formation of populations by fission that resulted in founder and drift effects have left their imprints on the genetic structures of contemporary populations".[27] The geneticist PP Majumder (2010) has recently argued that the findings of Reich et al. (2009) are in remarkable concordance with previous research using mtDNA and Y-DNA:[49]
Central Asian populations are supposed to have been major contributors to the Indian gene pool, particularly to the northern Indian gene pool, and the migrants had supposedly moved into India through what is now Afghanistan and Pakistan. Using mitochondrial DNA variation data collated from various studies, we have shown that populations of Central Asia and Pakistan show the lowest coefficient of genetic differentiation with the north Indian populations, a higher differentiation with the south Indian populations, and the highest with the northeast Indian populations. Northern Indian populations are genetically closer to Central Asians than populations of other geographical regions of India... . Consistent with the above findings, a recent study using over 500,000 biallelic autosomal markers has found a north to south gradient of genetic proximity of Indian populations to western Eurasians. This feature is likely related to the proportions of ancestry derived from the western Eurasian gene pool, which, as this study has shown, is greater in populations inhabiting northern India than those inhabiting southern India.
Genetic distance between caste groups and tribes
Studies by Watkins et al. (2005) and Kivisild et al. (2003) based on autosomal markers conclude that Indian caste and tribal populations have a common ancestry.[50][51] Reddy et al. (2005) found fairly uniform allele frequency distributions across caste groups of southern Andhra Pradesh, but significantly larger genetic distance between caste groups and tribes indicating genetic isolation of the tribes and castes.[52]
Viswanathan et al. (2004) in a study on genetic structure and affinities among tribal populations of southern India concludes, "Genetic differentiation was high and genetic distances were not significantly correlated with geographic distances. Genetic drift therefore probably played a significant role in shaping the patterns of genetic variation observed in southern Indian tribal populations. Otherwise, analyses of population relationships showed that all Indian and South Asian populations are still similar to one another, regardless of phenotypic characteristics, and do not show any particular affinities to Africans. We conclude that the phenotypic similarities of some Indian groups to Africans do not reflect a close relationship between these groups, but are better explained by convergence."[53]
A 2011 study published in the American Journal of Human Genetics[5] indicates that Indian ancestral components are the result of a more complex demographic history than was previously thought. According to the researchers, South Asia harbours two major ancestral components, one of which is spread at comparable frequency and genetic diversity in populations of Central Asia, West Asia and Europe; the other component is more restricted to South Asia. However, if one were to rule out the possibility of a large-scale Indo-Aryan migration, these findings suggest that the genetic affinities of both Indian ancestral components are the result of multiple gene flows over the course of thousands of years.[5]
See also
- Archaeogenetics
- Early human migrations
- Ethnic groups of South Asia
- Genetic history of indigenous peoples of the Americas
- Peopling of India
- Y-DNA haplogroups in populations of South Asia
- Genetic studies on Gujarati people
References
- 1 2 3 4 5 Kivisild, Toomas; et al. (1999), "The Place of the Indian Mitochondrial DNA Variants in the Global Network of Maternal Lineages and the Peopling of the Old World", Genomic Diversity: 135–152, ISBN 978-1-4613-6914-1, doi:10.1007/978-1-4615-4263-6_11
- 1 2 Baig, M. M.; Khan, A. A.; Kulkarni, K. M. (2004). "Mitochondrial DNA Diversity in Tribal and Caste Groups of Maharashtra (India) and its Implication on Their Genetic Origins". Annals of Human Genetics. 68 (5): 453–460. PMID 15469422. doi:10.1046/j.1529-8817.2004.00108.x.
- 1 2 Singh, Ashok Kumar (2007). Science & Technology For Upsc. Tata McGraw-Hill Education. p. 595. ISBN 978-0-07-065548-5.
- 1 2 3 4 Tripathy, Vikal; Nirmala, A.; Reddy, B. Mohan (2008), "Trends in Molecular Anthropological Studies in India" (PDF), International Journal of Human Genetics, 8 (1-2): 1–20
- 1 2 3 Metspalu, Mait; et al. (2011). "Shared and Unique Components of Human Population Structure and Genome-Wide Signals of Positive Selection in South Asia". The American Journal of Human Genetics. 89 (6): 731–44. PMC 3234374
 . PMID 22152676. doi:10.1016/j.ajhg.2011.11.010.
. PMID 22152676. doi:10.1016/j.ajhg.2011.11.010. - 1 2 Moorjani, Priya; et al. (2013). "Genetic Evidence for Recent Population Mixture in India". The American Journal of Human Genetics. 93 (3): 422–438. PMC 3769933
 . PMID 23932107. doi:10.1016/j.ajhg.2013.07.006.
. PMID 23932107. doi:10.1016/j.ajhg.2013.07.006. - 1 2 3 4 5 Kivisild, Toomas; et al. (2000), An Indian Ancestry: a Key for Understanding Human Diversity in Europe and Beyond (PDF), McDonald Institute Monographs
- 1 2 3 Y Haplogroups of the World, 2005, McDonald
- 1 2 3 4 5 6 7 8 9 10 11 12 Sengupta, Sanghamitra; et al. (2006). "Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists". The American Journal of Human Genetics. 78 (2): 202–21. PMC 1380230
 . PMID 16400607. doi:10.1086/499411.
. PMID 16400607. doi:10.1086/499411. - 1 2 3 4 5 6 7 8 Sahoo, S.; et al. (2006), "A prehistory of Indian Y chromosomes: Evaluating demic diffusion scenarios", Proceedings of the National Academy of Sciences, 103 (4): 843–8, Bibcode:2006PNAS..103..843S, PMC 1347984
 , PMID 16415161, doi:10.1073/pnas.0507714103
, PMID 16415161, doi:10.1073/pnas.0507714103 - 1 2 3 4 Thanseem, Ismail; et al. (2006). "Genetic affinities among the lower castes and tribal groups of India: Inference from Y chromosome and mitochondrial DNA". BMC Genetics. 7: 42. PMC 1569435
 . PMID 16893451. doi:10.1186/1471-2156-7-42.
. PMID 16893451. doi:10.1186/1471-2156-7-42. - 1 2 3 Zhao, Zhongming; et al. (2009). "Presence of three different paternal lineages among North Indians: A study of 560 Y chromosomes". Annals of Human Biology. 36 (1): 46–59. PMC 2755252
 . PMID 19058044. doi:10.1080/03014460802558522.
. PMID 19058044. doi:10.1080/03014460802558522. - ↑ Endicott, Metspalu & Kivisild 2007, p. 231.
- ↑ Endicott, Metspalu & Kivisild 2007, pp. 234-235.
- ↑ Oppenheimer 2003
- 1 2 Puente, Xoses; Velasco, Gloria; Gutiérrez-Fernández, Ana; Bertranpetit, Jaume; King, Mary-Claire; López-Otín, Carlos (2006). "Comparative analysis of cancer genes in the human and chimpanzee genomes". BMC Genomics. 7: 15. PMC 1382208
 . PMID 16438707. doi:10.1186/1471-2164-7-15.
. PMID 16438707. doi:10.1186/1471-2164-7-15. - 1 2 3 Metspalu, Mait; et al. (2004). "Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans". BMC Genetics. 5: 26. PMC 516768
 . PMID 15339343. doi:10.1186/1471-2156-5-26.
. PMID 15339343. doi:10.1186/1471-2156-5-26. - ↑ Kivisild, Toomas; et al. (1999a), "Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages" (PDF), Curr Biol, 9 (22): 1331–1334, PMID 10574762, doi:10.1016/s0960-9822(00)80057-3
- 1 2 Y-DNA Haplogroup H and its Subclades – 2015
- 1 2 3 4 5 Cordaux, Richard; et al. (2004). "Independent Origins of Indian Caste and Tribal Paternal Lineages". Current Biology. 14 (3): 231–5. PMID 14761656. doi:10.1016/j.cub.2004.01.024.
- 1 2 Thangaraj, Kumarasamy; et al. (2010). Cordaux, Richard, ed. "The Influence of Natural Barriers in Shaping the Genetic Structure of Maharashtra Populations". PLoS ONE. 5 (12): e15283. Bibcode:2010PLoSO...515283T. PMC 3004917
 . PMID 21187967. doi:10.1371/journal.pone.0015283.
. PMID 21187967. doi:10.1371/journal.pone.0015283. - ↑ Arunkumar, Ganeshprasad; Soria-Hernanz, David F.; Kavitha, Valampuri John; Arun, Varatharajan Santhakumari; Syama, Adhikarla; Ashokan, Kumaran Samy; Gandhirajan, Kavandanpatti Thangaraj; Vijayakumar, Koothapuli; Narayanan, Muthuswamy; Jayalakshmi, Mariakuttikan; Ziegle, Janet S.; Royyuru, Ajay K.; Parida, Laxmi; Wells, R. Spencer; Renfrew, Colin; Schurr, Theodore G.; Smith, Chris Tyler; Platt, Daniel E.; Pitchappan, Ramasamy (2012). "Population Differentiation of Southern Indian Male Lineages Correlates with Agricultural Expansions Predating the Caste System". PLoS ONE. 7 (11): e50269. Bibcode:2012PLoSO...750269A. PMC 3508930
 . PMID 23209694. doi:10.1371/journal.pone.0050269.
. PMID 23209694. doi:10.1371/journal.pone.0050269. - ↑ Sharma, S; Rai, E; Sharma, P; et al. (January 2009). "The Indian origin of paternal haplogroup R1a1* substantiates the autochthonous origin of Brahmins and the caste system". J. Hum. Genet. 54 (1): 47–55. PMID 19158816. doi:10.1038/jhg.2008.2.
- 1 2 Qamar, R; Ayub, Q; Mohyuddin, A; et al. (May 2002). "Y-Chromosomal DNA Variation in Pakistan". Am. J. Hum. Genet. 70: 1107–24. PMC 447589
 . PMID 11898125. doi:10.1086/339929.
. PMID 11898125. doi:10.1086/339929. - ↑ Shah AM, Tamang R, Moorjani P, Rani DS, Govindaraj P, Kulkarni G, Bhattacharya T, Mustak MS, Bhaskar LV, Reddy AG, Gadhvi D, Gai PB, Chaubey G, Patterson N, Reich D, Tyler-Smith C, Singh L, Thangaraj K (2011). "Indian Siddis: African Descendants with Indian Admixture". Am. J. Hum. Genet. 89: 154–61. PMC 3135801
 . PMID 21741027. doi:10.1016/j.ajhg.2011.05.030.
. PMID 21741027. doi:10.1016/j.ajhg.2011.05.030. - ↑ "The Genetics of Language and Farming Spread in India" (PDF).
- 1 2 3 4 Basu, A.; et al. (2003), "Ethnic India: A Genomic View, with Special Reference to Peopling and Structure", Genome Research, 13 (10): 2277–90, PMC 403703
 , PMID 14525929, doi:10.1101/gr.1413403
, PMID 14525929, doi:10.1101/gr.1413403 - ↑ Sengupta, S; Zhivotovsky, LA; King, R; et al. (February 2006). "Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists". Am. J. Hum. Genet. 78 (2): 202–21. PMC 1380230
 . PMID 16400607. doi:10.1086/499411.
. PMID 16400607. doi:10.1086/499411. - 1 2 3 4 5 Firasat, Sadaf; et al. (2006). "Y-chromosomal evidence for a limited Greek contribution to the Pathan population of Pakistan". European Journal of Human Genetics. 15 (1): 121–6. PMC 2588664
 . PMID 17047675. doi:10.1038/sj.ejhg.5201726.
. PMID 17047675. doi:10.1038/sj.ejhg.5201726. - 1 2 3 4 Sengupta et al. (2005)
- 1 2 3 4 Underhill, Peter A; et al. (2009), "Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a", European Journal of Human Genetics, 18 (4): 479–84, PMC 2987245
 , PMID 19888303, doi:10.1038/ejhg.2009.194
, PMID 19888303, doi:10.1038/ejhg.2009.194 - 1 2 3 4 5 6 7 Kivisild et al. (2003)
- 1 2 3 4 Sharma, Swarkar; et al. (2009). "The Indian origin of paternal haplogroup R1a1* substantiates the autochthonous origin of Brahmins and the caste system". Journal of Human Genetics. 54 (1): 47–55. PMID 19158816. doi:10.1038/jhg.2008.2.
- ↑ Mirabal, Sheyla; et al. (2009). "Y-Chromosome distribution within the geo-linguistic landscape of northwestern Russia". European Journal of Human Genetics. 17 (10): 1260–73. PMC 2986641
 . PMID 19259129. doi:10.1038/ejhg.2009.6.
. PMID 19259129. doi:10.1038/ejhg.2009.6. - 1 2 3 Underhill 2014.
- 1 2 3 Pamjav 2012.
- ↑ Fornarino et al. (2009)
- ↑ Manoukian, Jean-Grégoire (2006), "A Synthesis of Haplogroup R2 – 2006."
- 1 2 Kumar, Vikrant; et al. (2007). "Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic populations". BMC Evolutionary Biology. 7: 47. PMC 1851701
 . PMID 17389048. doi:10.1186/1471-2148-7-47.
. PMID 17389048. doi:10.1186/1471-2148-7-47. - ↑ Eaaswarkhanth, Muthukrishnan; et al. (2009). "Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations". European Journal of Human Genetics. 18 (3): 354–63. PMC 2859343
 . PMID 19809480. doi:10.1038/ejhg.2009.168.
. PMID 19809480. doi:10.1038/ejhg.2009.168. - ↑ The Place of the Indian mtDNA Variants in the Global Network of Maternal Lineages and the Peopling of the Old World
- ↑ "Ethnologue report for Indo-European". Ethnologue.com.
- ↑ Baldi, Philip (1990). Linguistic Change and Reconstruction Methodology. Walter de Gruyter. p. 342. ISBN 3-11-011908-0.
- ↑ Burling (2003), pp. 174–178.
- ↑ Bradley (2012) notes, MK in the wider sense including the Munda languages of eastern South Asia is also known as Austroasiatic.Languages and Language Families in China
- ↑ Bamshad, M; et al. (2001), "Genetic evidence on the origins of Indian caste populations", Genome Research, 11 (6): 994–1004, PMC 311057
 , PMID 11381027, doi:10.1101/gr.GR-1733RR
, PMID 11381027, doi:10.1101/gr.GR-1733RR - ↑ Mukherjee, Namita; et al. (2001), "High-resolution analysis of Y-chromosomal polymorphisms reveals signatures of population movements from central Asia and West Asia into India", Journal of Genetics, 80 (3): 125–35, PMID 11988631, doi:10.1007/BF02717908
- ↑ Reich, David; Thangaraj, Kumarasamy; Patterson, Nick; Price, Alkes L.; Singh, Lalji (2009). "Reconstructing Indian population history". Nature. 461 (7263): 489–94. Bibcode:2009Natur.461..489R. PMC 2842210
 . PMID 19779445. doi:10.1038/nature08365.
. PMID 19779445. doi:10.1038/nature08365. - ↑ Majumder, Partha P. (2010). "The Human Genetic History of South Asia". Current Biology. 20 (4): R184–7. PMID 20178765. doi:10.1016/j.cub.2009.11.053.
- ↑ Kivisild, T.; et al. (2003), "The Genetic Heritage of the Earliest Settlers Persists Both in Indian Tribal and Caste Populations", The American Journal of Human Genetics, 72 (2): 313–32, PMC 379225
 , PMID 12536373, doi:10.1086/346068
, PMID 12536373, doi:10.1086/346068 - ↑ Watkins, W.S.; et al. (2005). "Diversity and Divergence Among the Tribal Populations of India". Annals of Human Genetics. 69 (6): 680–692. doi:10.1046/j.1529-8817.2005.00200.x.
- ↑ Reddy, B. Mohan; et al. (2005). "Microsatellite Diversity in Andhra Pradesh, India: Genetic Stratification Versus Social Stratification". Human Biology. 77 (6): 803–23. PMID 16715839. doi:10.1353/hub.2006.0018.
- ↑ Vishwanathan, H.; et al. (2004). "Genetic structure and affinities among tribal populations of southern India: A study of 24 autosomal DNA markers". Annals of Human Genetics. 68 (2): 128–138. doi:10.1046/j.1529-8817.2003.00083.x.
- Additional references
- Allikas, Aire; et al. (2001), "Roles of the hinge region and the DNA binding domain of the bovine papillomavirus type 1 E2 protein in initiation of DNA replication", Virus Research, 75 (2): 95–106, PMID 11325464, doi:10.1016/S0168-1702(01)00219-2
- Behar, Doron M.; et al. (2004), "Contrasting patterns of Y chromosome variation in Ashkenazi Jewish and host non-Jewish European populations", Human Genetics, 114 (4): 354–65, PMID 14740294, doi:10.1007/s00439-003-1073-7
- Bhattacharyya, NP; et al. (1999), "Negligible male gene flow across ethnic boundaries in India, revealed by analysis of Y-chromosomal DNA polymorphisms", Genome Research, 9 (8): 711–9, PMID 10447506, doi:10.1101/gr.9.8.711 (inactive 2017-01-16)
- Cann, R. L. (2001), "Genetic Clues to Dispersal in Human Populations: Retracing the Past from the Present", Science, 291 (5509): 1742–8, Bibcode:2001Sci...291.1742C, PMID 11249820, doi:10.1126/science.1058948
- Cinnioglu, Cengiz; et al. (2004), "Excavating Y-chromosome haplotype strata in Anatolia", Human Genetics, 114 (2): 127–48, PMID 14586639, doi:10.1007/s00439-003-1031-4
- Das, Birajalaxmi; et al. (2004), "Minimal Sharing of Y-Chromosome STR Haplotypes Among Five Endogamous Population Groups from Western and Southwestern India", Human Biology, 76 (5): 743–63, PMID 15757245, doi:10.1353/hub.2005.0003
- Hemphill, Brian E.; Christensen, Alexander F. (3 November 1994). The Oxus Civilization as a Link between East and West: A Non-Metric Analysis of Bronze Age Bactrain Biological Affinities. Madison, Wisconsin. p. 13. (paper read at the South Asia Conference)
- Jobling, Mark A.; Tyler-Smith, Chris (2003), "The human Y chromosome: An evolutionary marker comes of age", Nature Reviews Genetics, 4 (8): 598–612, PMID 12897772, doi:10.1038/nrg1124
- Metspalu, Mait; et al. (2004), "Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans", BMC Genetics, 5: 26, PMC 516768
 , PMID 15339343, doi:10.1186/1471-2156-5-26
, PMID 15339343, doi:10.1186/1471-2156-5-26 - Patowary, Ashok; et al. (2012), "Systematic analysis and functional annotation of variations in the genome of an Indian individual", Human Mutation, 33 (7): 1133–40, PMID 22461382, doi:10.1002/humu.22091
- Rootsi, Siiri; et al. (2004), "Phylogeography of Y-Chromosome Haplogroup I Reveals Distinct Domains of Prehistoric Gene Flow in Europe", The American Journal of Human Genetics, 75 (1): 128–37, PMC 1181996
 , PMID 15162323, doi:10.1086/422196
, PMID 15162323, doi:10.1086/422196 - Qamar, Raheel; et al. (2002), "Y-Chromosomal DNA Variation in Pakistan", The American Journal of Human Genetics, 70 (5): 1107–24, PMC 447589
 , PMID 11898125, doi:10.1086/339929
, PMID 11898125, doi:10.1086/339929 - Semino, Ornella; et al. (2004), "Origin, Diffusion, and Differentiation of Y-Chromosome Haplogroups E and J: Inferences on the Neolithization of Europe and Later Migratory Events in the Mediterranean Area", The American Journal of Human Genetics, 74 (5): 1023–34, PMC 1181965
 , PMID 15069642, doi:10.1086/386295
, PMID 15069642, doi:10.1086/386295
Bibliography
- Indian Genome Variation Consortium (2008), "Genetic landscape of the people of India: A canvas for disease gene exploration", Journal of Genetics, 87 (1): 3–20, PMID 18560169, doi:10.1007/s12041-008-0002-x
- Endicott, Phillip; Metspalu, Mait; Kivisild, Toomas (2007), "Genetic evidence on modern human dispersals in South Asia: Y chromose and mitochondrial DNA perspectives", in Michael D. Petraglia; Bridget Allchin, The Evolution and History of Human Populations in South Asia, Springer, pp. 201–228, ISBN 1-4020-5561-7
- Hemphill, B.E.; Lukacs, J.R.; Kennedy, K.A.R. (1991). "Biological Adaptations and Affinities of Bronze Age Harappans". In Meadow, Richard H. Harappa excavations 1986–1990: a multidisciplinary approach to third millennium urbanism. pp. 137–82. ISBN 978-0-9629110-1-9.
- Kennedy, Kenneth A.R. (1984). "A Reassessment of the Theories of Racial Origins of the People of the Indus Valley Civilization from Recent Anthropological Data". In Kennedy, Kenneth A.R.; Possehl, Gregory L. Studies in the Archaeology and Palaeoanthropology of South Asia. Atlantic Highlands, NJ: Humanities Press. pp. 99–107.
- Kennedy, Kenneth A. R. (1995). "Have Aryans been identified in the prehistoric skeletal record from South Asia?". In George Erdosy. The Indo-Aryans of Ancient South Asia. Walter de Gruyter. pp. 49–54. ISBN 978-3-11-014447-5.
- Kivisild, Toomas (2000b). The origins of southern and western Eurasian populations: an mtDNA study (PDF). Tartu University, Estonia. (PhD)
- Kivisild, Toomas; et al. (2003a). "The Genetics of Language and Farming Spread in India". In Bellwood P, Renfrew C. Examining the farming/language dispersal hypothesis (PDF). McDonald Institute for Archaeological Research, Cambridge, United Kingdom. pp. 215–222.
- Mascarenhas, Desmond D.; Raina, Anupuma; Aston, Christopher E.; Sanghera, Dharambir K. (2015), "Genetic and Cultural Reconstruction of the Migration of an Ancient Lineage", BioMed Research International, 2015: 1–16, doi:10.1155/2015/651415
- Oppenheimer, Stephen (2003). The Real Eve: Modern Man's Journey out of Africa. New York: Carroll and Graf Publishers. ISBN 978-0-7867-1192-5.
- Pamjav (December 2012), "Brief communication: New Y-chromosome binary markers improve phylogenetic resolution within haplogroup R1a1", American Journal of Physical Anthropology, 149 (4): 611–615, PMID 23115110, doi:10.1002/ajpa.22167
- Renfrew, Colin; Boyle, Katie, eds. (2000a). An Indian Ancestry: a Key for Understanding Human Diversity in Europe and Beyond (PDF). ISBN 1-902937-08-2.
- Underhill, P. A. (2003), "Inferring Human History: Clues from Y-Chromosome Haplotypes", Cold Springer Harbor Symposia on Quantitative Biology, LXVIII: 487–493, PMID 15338652, doi:10.1101/sqb.2003.68.487
- Underhill, Peter A.; et al. (2015), "The phylogenetic and geographic structure of Y-chromosome haplogroup R1a" (PDF), European Journal of Human Genetics, 23 (1): 124–131, PMC 4266736
 , PMID 24667786, doi:10.1038/ejhg.2014.50
, PMID 24667786, doi:10.1038/ejhg.2014.50 - Wells, S (2003). The Journey of Man: A Genetic Odyssey. Princeton University Press.
External links
- Introduction to Haplogroups and Haplotypes, Mark A. Jobling, University of Leicester.
- Journey of Man: Peopling of the World, Bradshaw Foundation, in association with Stephen Oppenheimer.
- Indian Genome Variation Database Institute of Genomics and Integrative Biology
- Indian Genome Variation Consortium (2008). "Genetic landscape of the people of India: a canvas for disease gene exploration". Journal of Genetics. 87 (1): 3–20. PMID 18560169. doi:10.1007/s12041-008-0002-x.
- List of R2 frequency
- Maldives