Glyceraldehyde 3-phosphate dehydrogenase
| Glyceraldehyde 3-phosphate dehydrogenase, NAD binding domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 determinants of enzyme thermostability observed in the molecular structure of thermus aquaticus d-glyceraldehyde-3-phosphate dehydrogenase at 2.5 angstroms resolution | |||||||||
| Identifiers | |||||||||
| Symbol | Gp_dh_N | ||||||||
| Pfam | PF00044 | ||||||||
| Pfam clan | CL0063 | ||||||||
| InterPro | IPR020828 | ||||||||
| PROSITE | PDOC00069 | ||||||||
| SCOP | 1gd1 | ||||||||
| SUPERFAMILY | 1gd1 | ||||||||
| |||||||||
| Glyceraldehyde 3-phosphate dehydrogenase, C-terminal domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 crystal structure of glyceraldehyde-3-phosphate dehydrogenase from pyrococcus horikoshii ot3 | |||||||||
| Identifiers | |||||||||
| Symbol | Gp_dh_C | ||||||||
| Pfam | PF02800 | ||||||||
| Pfam clan | CL0139 | ||||||||
| InterPro | IPR020829 | ||||||||
| PROSITE | PDOC00069 | ||||||||
| SCOP | 1gd1 | ||||||||
| SUPERFAMILY | 1gd1 | ||||||||
| |||||||||
Glyceraldehyde 3-phosphate dehydrogenase (abbreviated as GAPDH or less commonly as G3PDH) (EC 1.2.1.12) is an enzyme of ~37kDa that catalyzes the sixth step of glycolysis and thus serves to break down glucose for energy and carbon molecules. In addition to this long established metabolic function, GAPDH has recently been implicated in several non-metabolic processes, including transcription activation, initiation of apoptosis,[3] ER to Golgi vesicle shuttling, and fast axonal, or axoplasmic transport.[4] In sperm, a testis-specific isoenzyme GAPDHS is expressed.
Structure
Under normal cellular conditions, cytoplasmic GAPDH exists primarily as a tetramer. This form is composed of four identical 37-kDa subunits containing a single catalytic thiol group each and critical to the enzyme's catalytic function.[5][6] Nuclear GAPDH has increased isoelectric point (pI) of pH 8.3–8.7.[6] Of note, the cysteine residue C152 in the enzyme's active site is required for the induction of apoptosis by oxidative stress.[6] Notably, post-translational modifications of cytoplasmic GAPDH contribute to its functions outside of glycolysis.[5]
Interestingly, GAPDH is encoded by a single gene that produces a single mRNA transcript with no known splice variants, though an isoform does exist as a separate gene that is expressed only in spermatozoa.[6]
Reaction
| glyceraldehyde 3-phosphate | glyceraldehyde phosphate dehydrogenase | D-glycerate 1,3-bisphosphate | |
 |
 | ||
| NAD+ +Pi | NADH + H+ | ||
 | |||
| NAD+ +Pi | NADH + H+ | ||
Compound C00118 at KEGG Pathway Database. Enzyme 1.2.1.12 at KEGG Pathway Database. Reaction R01063 at KEGG Pathway Database. Compound C00236 at KEGG Pathway Database.
Two-step conversion of G3P
The first reaction is the oxidation of glyceraldehyde 3-phosphate (G3P) at the position-1 (in the diagram it is shown as the 4th carbon from glycolysis), in which an aldehyde is converted into a carboxylic acid (ΔG°'=-50 kJ/mol (−12kcal/mol)) and NAD+ is simultaneously reduced endergonically to NADH.
The energy released by this highly exergonic oxidation reaction drives the endergonic second reaction (ΔG°'=+50 kJ/mol (+12kcal/mol)), in which a molecule of inorganic phosphate is transferred to the GAP intermediate to form a product with high phosphoryl-transfer potential: 1,3-bisphosphoglycerate (1,3-BPG).
This is an example of phosphorylation coupled to oxidation, and the overall reaction is somewhat endergonic (ΔG°'=+6.3 kJ/mol (+1.5)). Energy coupling here is made possible by GAPDH.
Mechanism
GAPDH uses covalent catalysis and general base catalysis to decrease the very large and positive activation energy of the second step of this reaction. First, a cysteine residue in the active site of GAPDH attacks the carbonyl group of GAP, creating a hemithioacetal intermediate (covalent catalysis). Next, an adjacent, tightly bound molecule of NAD+ accepts a hydride ion from GAP, forming NADH; GAP is concomitantly oxidized to a thioester intermediate using a molecule of water. This thioester species is much higher in energy than the carboxylic acid species that would result in the absence of GAPDH (the carboxylic acid species is so low in energy that the energy barrier for the second step of the reaction (phosphorylation) would be too high, and the reaction, therefore, too slow and equilibrium too unfavorable for a living organism). Donation of the hydride ion by the hemithioacetal is facilitated by its deprotonation by a histidine residue in the enzyme's active site (general base catalysis). Deprotonation encourages the reformation of the carbonyl group in the thioester intermediate and ejection of the hydride ion. NADH leaves the active site and is replaced by another molecule of NAD+, the positive charge of which stabilizes the negatively charged carbonyl oxygen in the transition state of the next and ultimate step. Finally, a molecule of inorganic phosphate attacks the thioester and forms a tetrahedral intermediate, which then collapses to release 1,3-bisphosphoglycerate, and the thiol group of the enzyme's cysteine residue.
Regulation
This protein may use the morpheein model of allosteric regulation.[7]
Function
Metabolic
As its name indicates, glyceraldehyde 3-phosphate dehydrogenase (GAPDH) catalyses the conversion of glyceraldehyde 3-phosphate to D-glycerate 1,3-bisphosphate. This is the 6th step in the glycolytic breakdown of glucose, an important pathway of energy and carbon molecule supply which takes place in the cytosol of eukaryotic cells. The conversion occurs in two coupled steps. The first is favourable and allows the second unfavourable step to occur.
Transcription and apoptosis
GAPDH can itself activate transcription. The OCA-S transcriptional coactivator complex contains GAPDH and lactate dehydrogenase, two proteins previously only thought to be involved in metabolism. GAPDH moves between the cytosol and the nucleus and may thus link the metabolic state to gene transcription.[8]
In 2005, Hara et al. showed that GAPDH initiates apoptosis. This is not a third function, but can be seen as an activity mediated by GAPDH binding to DNA like in transcription activation, discussed above. The study demonstrated that GAPDH is S-nitrosylated by NO in response to cell stress, which causes it to bind to the protein SIAH1, a ubiquitin ligase. The complex moves into the nucleus where Siah1 targets nuclear proteins for degradation, thus initiating controlled cell shutdown.[9] In subsequent study the group demonstrated that deprenyl, which has been used clinically to treat Parkinson's disease, strongly reduces the apoptotic action of GAPDH by preventing its S-nitrosylation and might thus be used as a drug.[10]
Metabolic switch
GAPDH acts as reversible metabolic switch under oxidative stress.[11] When cells are exposed to oxidants, they need excessive amounts of the antioxidant cofactor NADPH. In the cytosol, NADPH is reduced from NADP+ by several enzymes, three of them catalyze the first steps of the Pentose phosphate pathway. Oxidant-treatments cause an inactivation of GAPDH. This inactivation re-routes temporally the metabolic flux from glycolysis to the Pentose Phosphate Pathway, allowing the cell to generate more NADPH.[12] Under stress conditions, NADPH is needed by some antioxidant-systems including glutaredoxin and thioredoxin as well as being essential for the recycling of gluthathione.
ER to Golgi transport
GAPDH also appears to be involved in the vesicle transport from the endoplasmic reticulum (ER) to the Golgi apparatus which is part of shipping route for secreted proteins. It was found that GAPDH is recruited by rab2 to the vesicular-tubular clusters of the ER where it helps to form COP 1 vesicles. GAPDH is activated via tyrosine phosphorylation by Src.[13]
Additional functions
GAPDH, like many other enzymes, has multiple functions. In addition to catalysing the 6th step of glycolysis, recent evidence implicates GAPDH in other cellular processes.GAPDH has been described to exhibit higher order multifunctionality in the context of maintaining cellular iron homeostasis.[14] This came as a surprise to researchers but it makes evolutionary sense to re-use and adapt existing proteins instead of evolving a novel protein from scratch.
Use as loading control
Because the GAPDH gene is often stably and constitutively expressed at high levels in most tissues and cells, it is considered a housekeeping gene. For this reason, GAPDH is commonly used by biological researchers as a loading control for western blot and as a control for qPCR. However, researchers have reported different regulation of GAPDH under specific conditions.[15] For example, the transcription factor MZF-1 has been shown to regulate the GAPDH gene.[16] Therefore, the use of GAPDH as loading control has to be considered carefully.
Cellular distribution
All steps of glycolysis take place in the cytosol and so does the reaction catalysed by GAPDH. In red blood cells, GAPDH and several other glycolytic enzymes assemble in complexes on the inside of the cell membrane. The process appears to be regulated by phosphorylation and oxygenation.[17] Bringing several glycolytic enzymes close to each other is expected to greatly increase the overall speed of glucose breakdown. Recent studies have also revealed that GAPDH is expressed in an iron dependent fashion on the exterior of the cell membrane a where it plays a role in maintenance of cellular iron homeostasis.[18][19]
Clinical significance
Cancer
GAPDH is overexpressed in multiple human cancers, such as cutaneous melanoma, and its expression is positively correlated with tumor progression.[20][21] Its glycolytic and antiapoptotic functions contribute to proliferation and protection of tumor cells, promoting tumorigenesis. Notably, GAPDH protects against telomere shortening induced by chemotherapeutic drugs that stimulate the sphingolipid ceramide. Meanwhile, conditions like oxidative stress impair GAPDH function, leading to cellular aging and death.[6] Moreover, depletion of GAPDH has managed to induce senescence in tumor cells, thus presenting a novel therapeutic strategy for controlling tumor growth.[22]
Neurodegeneration
GAPDH has been implicated in several neurodegenerative diseases and disorders, largely through interactions with other proteins specific to that disease or disorder. These interactions may affect not only energy metabolism but also other GAPDH functions.[5] For example, GAPDH interactions with beta-amyloid precursor protein (betaAPP) could interfere with its function regarding the cytoskeleton or membrane transport, while interactions with huntingtin could interfere with its function regarding apoptosis, nuclear tRNA transport, DNA replication, and DNA repair. In addition, nuclear translocation of GAPDH has been reported in Parkinson's disease (PD), and several anti-apoptotic PD drugs, such as rasagiline, function by preventing the nuclear translocation of GAPDH. It is proposed that hypometabolism may be one contributor to PD, but the exact mechanisms underlying GAPDH involvement in neurodegenerative disease remains to be clarified.[23] The SNP rs3741916 in the 5' UTR of the GAPDH gene may be associated with late onset Alzheimer's disease.[24]
Interactions
Protein binding partners
GAPDH participates in a number of biological functions through its protein–protein interactions with:
- tubulin to facilitate microtubule bundling;[5]
- actin to facilitate actin polymerization;[5]
- VDAC1 to induce mitochondrial membrane permeabilization (MMP) and apoptosis;[5]
- Inositol 1,4,5-trisphosphate receptor to regulate intracellular Ca2+ signaling;[5]
- Oct-1 to form the coactivator complex OCA-S, which is required for histone H2B synthesis during S phase of the cell cycle;[6]
- p22 to aid microtubule organization;[6]
- Rab2 to facilitate endoplasmic reticulum (ER)–golgi transport;[6]
- Transferrin on macrophage cell surfaces;[6][19][25]
- Lactate dehydrogenase;[6]
- Lactoferrin;[26]
- Apurinic/apyrimidinic endonuclease (APE1), thus converting oxidized APE1 to its reduced form, to restart its endonuclease activity;[6]
- Promyelocytic leukaemia protein (PML) in an RNA-dependent fashion;[6]
- Rheb to sequester the GTPase during low glucose conditions;[6]
- Siah1 to form a complex that translocates to the nucleus, where it ubiquitinates and degrades nuclear proteins during nitrosative stress conditions;[6]
- GAPDH's competitor of Siah protein enhances life (GOSPEL) to block GAPDH interaction with Siah1 and, thus, cell death in response to oxidative stress;[6]
- p300/CREB binding protein (CBP), which acetylates GAPDH and, in turn, enhances the acetylation of additional apoptotic targets;[6]
- skeletal muscle-specific Ca2+/calmodulin-dependent protein kinase;[6]
- Akt;[6]
- Beta-amyloid precursor protein (betaAPP);[23]
- Huntingtin.[23]
- GAPDH can self-associate into homotypic oligomers/aggregates
Nucleic acid binding partners
GAPDH binds to single-stranded RNA and DNA and a number of nucleic acid binding partners have been identified:[6]
- tRNA,
- Hepatitis A viral RNA,
- Hepatitis B viral RNA,
- Hepatitis C viral RNA,
- HPIV3,
- lymphokine mRNA,
- IFN-γ mRNA,
- JEV mRNA, and
- telomeric DNA.
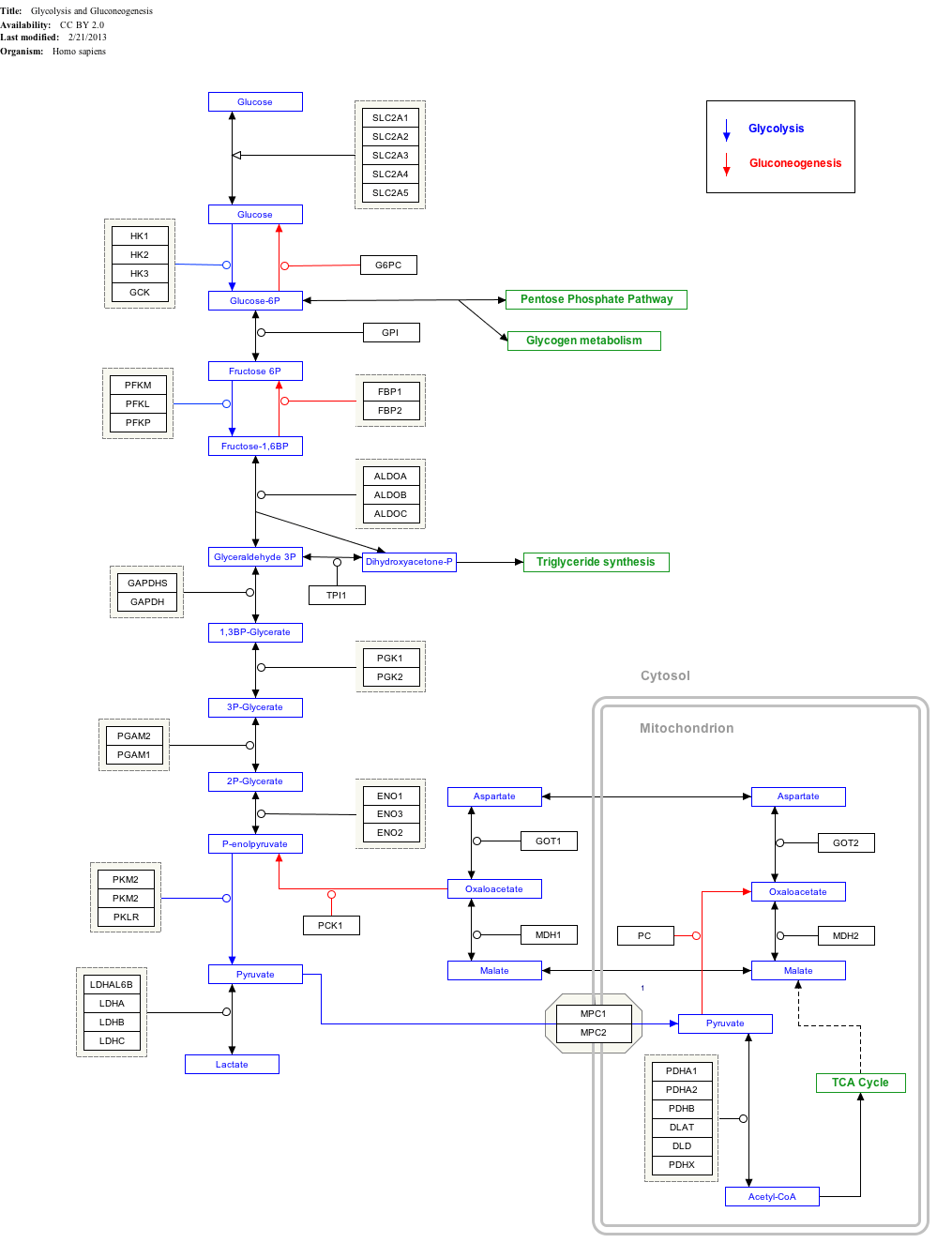
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
|{{{bSize}}}px|alt=Glycolysis and Gluconeogenesis edit]]
Glycolysis and Gluconeogenesis edit
- ↑ The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Tarze A, Deniaud A, Le Bras M, Maillier E, Molle D, Larochette N, Zamzami N, Jan G, Kroemer G, Brenner C (April 2007). "GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization". Oncogene. 26 (18): 2606–20. PMID 17072346. doi:10.1038/sj.onc.1210074.
- ↑ Zala D, Hinckelmann MV, Yu H, Lyra da Cunha MM, Liot G, Cordelières FP, Marco S, Saudou F (January 2013). "Vesicular glycolysis provides on-board energy for fast axonal transport". Cell. 152 (3): 479–91. PMID 23374344. doi:10.1016/j.cell.2012.12.029.
- 1 2 3 4 5 6 7 Tristan C, Shahani N, Sedlak TW, Sawa A (February 2011). "The diverse functions of GAPDH: views from different subcellular compartments". Cellular Signalling. 23 (2): 317–23. PMC 3084531
 . PMID 20727968. doi:10.1016/j.cellsig.2010.08.003.
. PMID 20727968. doi:10.1016/j.cellsig.2010.08.003. - 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 Nicholls C, Li H, Liu JP (August 2012). "GAPDH: a common enzyme with uncommon functions". Clinical and Experimental Pharmacology & Physiology. 39 (8): 674–9. PMID 21895736. doi:10.1111/j.1440-1681.2011.05599.x.
- ↑ Selwood T, Jaffe EK (March 2012). "Dynamic dissociating homo-oligomers and the control of protein function". Archives of Biochemistry and Biophysics. 519 (2): 131–43. PMC 3298769
 . PMID 22182754. doi:10.1016/j.abb.2011.11.020.
. PMID 22182754. doi:10.1016/j.abb.2011.11.020. - ↑ Zheng L, Roeder RG, Luo Y (July 2003). "S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component". Cell. 114 (2): 255–66. PMID 12887926. doi:10.1016/S0092-8674(03)00552-X.
- ↑ Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A (July 2005). "S-nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding". Nature Cell Biology. 7 (7): 665–74. PMID 15951807. doi:10.1038/ncb1268.
- ↑ Hara MR, Thomas B, Cascio MB, Bae BI, Hester LD, Dawson VL, Dawson TM, Sawa A, Snyder SH (March 2006). "Neuroprotection by pharmacologic blockade of the GAPDH death cascade". Proceedings of the National Academy of Sciences of the United States of America. 103 (10): 3887–9. PMC 1450161
 . PMID 16505364. doi:10.1073/pnas.0511321103.
. PMID 16505364. doi:10.1073/pnas.0511321103. - ↑ Agarwal AR, Zhao L, Sancheti H, Sundar IK, Rahman I, Cadenas E (November 2012). "Short-term cigarette smoke exposure induces reversible changes in energy metabolism and cellular redox status independent of inflammatory responses in mouse lungs". American Journal of Physiology. Lung Cellular and Molecular Physiology. 303 (10): L889–98. PMID 23064950. doi:10.1152/ajplung.00219.2012.
- ↑ Ralser M, Wamelink MM, Kowald A, Gerisch B, Heeren G, Struys EA, Klipp E, Jakobs C, Breitenbach M, Lehrach H, Krobitsch S (2007). "Dynamic rerouting of the carbohydrate flux is key to counteracting oxidative stress". Journal of Biology. 6 (4): 10. PMC 2373902
 . PMID 18154684. doi:10.1186/jbiol61.
. PMID 18154684. doi:10.1186/jbiol61. - ↑ Tisdale EJ, Artalejo CR (June 2007). "A GAPDH mutant defective in Src-dependent tyrosine phosphorylation impedes Rab2-mediated events". Traffic. 8 (6): 733–41. PMID 17488287. doi:10.1111/j.1600-0854.2007.00569.x.
- ↑ Boradia VM, Raje M, Raje CI (December 2014). "Protein moonlighting in iron metabolism: glyceraldehyde-3-phosphate dehydrogenase (GAPDH)". Biochemical Society Transactions. 42 (6): 1796–801. PMID 25399609. doi:10.1042/BST20140220.
- ↑ Barber RD, Harmer DW, Coleman RA, Clark BJ (May 2005). "GAPDH as a housekeeping gene: analysis of GAPDH mRNA expression in a panel of 72 human tissues". Physiological Genomics. 21 (3): 389–95. PMID 15769908. doi:10.1152/physiolgenomics.00025.2005.
- ↑ Piszczatowski RT, Rafferty BJ, Rozado A, Tobak S, Lents NH (August 2014). "The glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) is regulated by myeloid zinc finger 1 (MZF-1) and is induced by calcitriol". Biochemical and Biophysical Research Communications. 451 (1): 137–41. PMID 25065746. doi:10.1016/j.bbrc.2014.07.082.
- ↑ Campanella ME, Chu H, Low PS (February 2005). "Assembly and regulation of a glycolytic enzyme complex on the human erythrocyte membrane". Proceedings of the National Academy of Sciences of the United States of America. 102 (7): 2402–7. PMC 549020
 . PMID 15701694. doi:10.1073/pnas.0409741102.
. PMID 15701694. doi:10.1073/pnas.0409741102. - ↑ Sirover MA (December 2014). "Structural analysis of glyceraldehyde-3-phosphate dehydrogenase functional diversity". The International Journal of Biochemistry & Cell Biology. 57: 20–6. PMC 4268148
 . PMID 25286305. doi:10.1016/j.biocel.2014.09.026.
. PMID 25286305. doi:10.1016/j.biocel.2014.09.026. - 1 2 Kumar S, Sheokand N, Mhadeshwar MA, Raje CI, Raje M (January 2012). "Characterization of glyceraldehyde-3-phosphate dehydrogenase as a novel transferrin receptor". The International Journal of Biochemistry & Cell Biology. 44 (1): 189–99. PMID 22062951. doi:10.1016/j.biocel.2011.10.016.
- ↑ Ramos D, Pellín-Carcelén A, Agustí J, Murgui A, Jordá E, Pellín A, Monteagudo C (January 2015). "Deregulation of glyceraldehyde-3-phosphate dehydrogenase expression during tumor progression of human cutaneous melanoma". Anticancer Research. 35 (1): 439–44. PMID 25550585.
- ↑ Wang D, Moothart DR, Lowy DR, Qian X (2013). "The expression of glyceraldehyde-3-phosphate dehydrogenase associated cell cycle (GACC) genes correlates with cancer stage and poor survival in patients with solid tumors". PLOS ONE. 8 (4): e61262. PMC 3631177
 . PMID 23620736. doi:10.1371/journal.pone.0061262.
. PMID 23620736. doi:10.1371/journal.pone.0061262. - ↑ Phadke M, Krynetskaia N, Mishra A, Krynetskiy E (July 2011). "Accelerated cellular senescence phenotype of GAPDH-depleted human lung carcinoma cells". Biochemical and Biophysical Research Communications. 411 (2): 409–15. PMC 3154080
 . PMID 21749859. doi:10.1016/j.bbrc.2011.06.165.
. PMID 21749859. doi:10.1016/j.bbrc.2011.06.165. - 1 2 3 Mazzola JL, Sirover MA (October 2002). "Alteration of intracellular structure and function of glyceraldehyde-3-phosphate dehydrogenase: a common phenotype of neurodegenerative disorders?". Neurotoxicology. 23 (4–5): 603–9. PMID 12428732. doi:10.1016/s0161-813x(02)00062-1.
- ↑ Allen M, Cox C, Belbin O, Ma L, Bisceglio GD, Wilcox SL, Howell CC, Hunter TA, Culley O, Walker LP, Carrasquillo MM, Dickson DW, Petersen RC, Graff-Radford NR, Younkin SG, Ertekin-Taner N (January 2012). "Association and heterogeneity at the GAPDH locus in Alzheimer's disease". Neurobiology of Aging. 33 (1): 203.e25–33. PMC 3017231
 . PMID 20864222. doi:10.1016/j.neurobiolaging.2010.08.002.
. PMID 20864222. doi:10.1016/j.neurobiolaging.2010.08.002. - ↑ Raje CI, Kumar S, Harle A, Nanda JS, Raje M (February 2007). "The macrophage cell surface glyceraldehyde-3-phosphate dehydrogenase is a novel transferrin receptor". The Journal of Biological Chemistry. 282 (5): 3252–61. PMID 17121833. doi:10.1074/jbc.M608328200.
- ↑ Secreted multifunctional Glyceraldehyde-3-phosphate dehydrogenase sequesters lactoferrin and iron into cells via a non-canonical pathway. Anoop S. Chauhan, Pooja Rawat, Himanshu Malhotra, Navdeep Sheokand, Manoj Kumar, Anil Patidar, Surbhi Chaudhary, Priyanka Jakhar, Chaaya I. Raje and Manoj Raje Scientific Reports 5, 18465; doi:10.1038/srep18465 (2015)
Further reading
- Voet D, Voet JG (2010). Biochemistry. New York: Wiley. ISBN 0-470-57095-4.
- Stryer L, Berg JM, Tymoczko JL (2002). Biochemistry, Fifth Edition & Lecture Notebook. San Francisco: W. H. Freeman. ISBN 0-7167-9804-2.
- diagram of the GAPDH reaction mechanism from Lodish MCB at NCBI bookshelf
- similar diagram from Alberts The Cell at NCBI bookshelf
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||