Neomycin
 | |
 | |
| Clinical data | |
|---|---|
| Trade names | Neo-rx |
| AHFS/Drugs.com | Monograph |
| MedlinePlus | a682274 |
| Pregnancy category |
|
| Routes of administration | Topical, oral |
| ATC code | |
| Legal status | |
| Legal status | |
| Pharmacokinetic data | |
| Bioavailability | None |
| Protein binding | N/A |
| Metabolism | N/A |
| Biological half-life | 2 to 3 hours |
| Identifiers | |
| |
| CAS Number | |
| PubChem CID | |
| IUPHAR/BPS | |
| DrugBank | |
| ChemSpider | |
| UNII | |
| KEGG | |
| ChEBI | |
| ChEMBL | |
| ECHA InfoCard | 100.014.333 |
| Chemical and physical data | |
| Formula | C23H46N6O13 |
| Molar mass | 614.644 g/mol |
| 3D model (JSmol) | |
| |
| |
| | |
Neomycin is an aminoglycoside antibiotic found in many topical medications such as creams, ointments, and eyedrops. The discovery of neomycin dates back to 1949. It was discovered in the lab of Selman Waksman. Neomycin belongs to aminoglycoside class of antibiotics that contain two or more aminosugars connected by glycosidic bonds.
Uses
Neomycin is typically used as a topical preparation, such as Neosporin. It can also be given orally, where it is usually combined with other antibiotics. Neomycin is not absorbed from the gastrointestinal tract and has been used as a preventive measure for hepatic encephalopathy and hypercholesterolemia. By killing bacteria in the intestinal tract, it keeps ammonia levels low and prevents hepatic encephalopathy, especially prior to GI surgery. It has also been used to treat small intestinal bacterial overgrowth. It is not given via injection, as neomycin is extremely nephrotoxic (causes kidney damage), even when compared to other aminoglycosides. The exception is when neomycin is included, in very small quantities, as a preservative in some vaccines – typically 25 mg per dose.[1]
Molecular biology
Neomycin resistance is conferred by either one of two aminoglycoside phosphotransferase genes.[2] A neo gene is commonly included in DNA plasmids used by molecular biologists to establish stable mammalian cell lines expressing cloned proteins in culture; many commercially available protein expression plasmids contain neo as a selectable marker. Non-transfected cells will eventually die off when the culture is treated with neomycin or similar antibiotic. Neomycin or kanamycin can be used for prokaryotes, but geneticin (G418) is, in general, needed for eukaryotes.
Neomycin binds to the 30S subunit of the ribosome and inhibits translation of proteins from mRNA.[3]
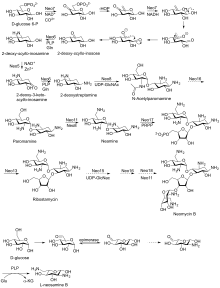
Biosynthetic pathway
First isolated from the Streptomyces fradiae and Streptomyces albogriseus in 1949 (NBRC 12773) [4]. Neomycin is a mixture of neomycin B (framycetin); and its epimer neomycin C, the latter component accounting for some 5–15% of the mixture. Neomycin has good activity against Gram-positive and Gram-negative bacteria, but is very ototoxic. Its use is thus restricted to oral treatment of intestinal infections.[5]
Neomycin B is composed of four parts: D-neosamine, 2-deoxystreptamine (2-DOS), D-ribose, and L-neosamine.
Neomycin A, also called neamine, contains D-neosamine and 2-deoxystreptamine. Neamine is made from six genes, DOIS gene (btrC, neo7); L-glutamine:DOI aminotransferase gene (btrS, neo6); a putative glycosyltransferase gene (btrM, neo8); a putative aminotransferase (similar to glutamate-1-semialdehyde 2,1- aminomutase) gene (btrB, neo18); a putative alcohol dehydrogenase gene (btrE, neo5); another putative dehydrogenase (similar to chorine dehydrogenase and related flavoproteins) gene (btrQ, neo11).[6] A deacetylase acting to remove the acetyl group on N-acetylglucosamine moieties of aminoglycoside intermediates (Neo16), still needs to be clarified (sequence similar to BtrD).[7]
Next is the attachment of the D-ribose via ribosylation of neamine, using 5-phosphoribosyl-1-diphosphate (PRPP) as the ribosyl donor (BtrL, BtrP);[8] glycosyltransferase (potential homologues RibF, LivF, Parf) gene (Neo15).[9]
Neosamine B (L-neosamine B) is most likely biosynthesized in the same manner as the neosamine C (D-niosamine) in neamine biosynthesis, but with an additional epimerization step required to account for the presence of the epimeric neosamine B in neomycin B.[10]
Spectrum
Similar to other aminoglycosides, neomycin has excellent activity against Gram-negative bacteria, and has partial activity against Gram-positive bacteria. It is relatively toxic to humans, and many people have allergic reactions to it.[11] See: Hypersensitivity. Physicians sometimes recommend using antibiotic ointments without neomycin, such as Polysporin.[12] The following represents MIC susceptibility data for a few medically significant Gram-negative bacteria.[13]
- Enterobacter cloacae: >16 μg/ml
- Escherichia coli: 1 μg/ml
- Proteus vulgaris: 0.25 μg/ml
Composition
Standard grade neomycin is composed of a number of related compounds including neomycin A (neamine), neomycin B (framycetin), neomycin C, and a few minor compounds found in much lower quantities. Neomycin B is the most active component in neomycin followed by neomycin C and neomycin A. Neomycin A is an inactive degradation product of the C and B isomers.[14] The quantities of these components in neomycin vary from lot-to-lot depending on the manufacturer and manufacturing process.[15]
Safety
In 2005–06, neomycin was the fifth-most-prevalent allergen in patch test results (10.0%).[16] Neomycin, like other aminoglycosides, has been shown to be ototoxic causing tinnitus, hearing loss, and vestibular problems in a small number of patients. Patients with existing tinnitus or sensorineural hearing loss should speak with their doctor about the risks and side effects prior to taking this medication.
History
Neomycin was discovered in 1949 by the microbiologist Selman Waksman and his student Hubert Lechevalier at Rutgers University. It is produced naturally by the bacterium Streptomyces fradiae.[17] Synthesis requires specific nutrient conditions in either stationary or submerged aerobic conditions. The compound is then isolated and purified from the bacterium.[18]
DNA binding
Aminoglycosides such as neomycin are known for their ability to bind to duplex RNA with high affinity.[19] The association constant for neomycin with A-site RNA has been found to be in the 109 M−1 range.[20] However, more than 50 years after its discovery, its DNA-binding properties were still unknown. Neomycin has been shown to induce thermal stabilization of triplex DNA, while having little or almost no effect on the B-DNA duplex stabilization.[21] Neomycin was also shown to bind to structures that adopt A-form structure, triplex DNA being one of them. Neomycin also includes DNA:RNA hybrid triplex formation.[22]
References
- ↑ Heidary, Noushin; Cohen, David E. (2005). "Hypersensitivity reactions to vaccine components". Dermatitis. 16 (3): 115–20. PMID 16242081. doi:10.1097/01206501-200509000-00004.
- ↑ "G418/neomycin-cross resistance?". Retrieved 2008-10-19.
- ↑ Mehta, Roopal; Champney, W. Scott (2003). "Neomycin and Paromomycin Inhibit 30S Ribosomal Subunit Assembly in Staphylococcus aureus". Current Microbiology. 47 (3): 237–43. PMID 14570276. doi:10.1007/s00284-002-3945-9.
- ↑ Waksman, Selman A.; Lechevalier, Hubert A.; Harris, Dale A. (1949). "Neomycin—Production and Antibiotic Properties 123". Journal of Clinical Investigation. 28 (5 Pt 1): 934–9. PMC 438928
 . PMID 16695766. doi:10.1172/JCI102182.
. PMID 16695766. doi:10.1172/JCI102182. - ↑ Dewick M., Dewick (March 2009). Dr. (3rd ed.). The Atrium, Southern Gate, Chichester, West Sussex, PO19 8SQ, United Kingdom (Page 4).: John Wiley and Sons Ltd. pp. 508, 510, 511. ISBN 978-0-470-74168-9.
- ↑ Kudo, Fumitaka; Yamamoto, Yasuhito; Yokoyama, Kenichi; Eguchi, Tadashi; Kakinuma, Katsumi (2005). "Biosynthesis of 2-Deoxystreptamine by Three Crucial Enzymes in Streptomyces fradiae NBRC 12773". The Journal of Antibiotics. 58 (12): 766–74. PMID 16506694. doi:10.1038/ja.2005.104.
- ↑ Park, Je Won; Park, Sung Ryeol; Nepal, Keshav Kumar; Han, Ah Reum; Ban, Yeon Hee; Yoo, Young Ji; Kim, Eun Ji; Kim, Eui Min; Kim, Dooil; Sohng, Jae Kyung; Yoon, Yeo Joon (2011). "Discovery of parallel pathways of kanamycin biosynthesis allows antibiotic manipulation". Nature Chemical Biology. 7 (11): 843–52. PMID 21983602. doi:10.1038/nchembio.671.
- ↑ Kudo, Fumitaka; Fujii, Takuya; Kinoshita, Shunsuke; Eguchi, Tadashi (2007). "Unique O-ribosylation in the biosynthesis of butirosin". Bioorganic & Medicinal Chemistry. 15 (13): 4360–8. PMID 17482823. doi:10.1016/j.bmc.2007.04.040.
- ↑ Fan, Qingzhi; Huang, Fanglu; Leadlay, Peter F.; Spencer, Jonathan B. (2008). "The neomycin biosynthetic gene cluster of Streptomyces fradiae NCIMB 8233: Genetic and biochemical evidence for the roles of two glycosyltransferases and a deacetylase". Organic & Biomolecular Chemistry. 6 (18): 3306–14. PMID 18802637. doi:10.1039/B808734B.
- ↑ Llewellyn, Nicholas M.; Spencer, Jonathan B. (2006). "Biosynthesis of 2-deoxystreptamine-containing aminoglycoside antibiotics". Natural Product Reports. 23 (6): 864–74. PMID 17119636. doi:10.1039/B604709M.
- ↑ DermNet dermatitis/neomycin-allergy
- ↑ "Your Medicine Cabinet". DERMAdoctor.com, Inc. Retrieved 2008-10-19.
- ↑ "Neomycin sulfate, EP Susceptibility and Minimum Inhibitory Concentration (MIC) Data" (PDF). TOKU-E.
- ↑ Cammack, R. Attwood, T. K. Campbell, P. N. Parish, J. H. Smith, A. D. Stirling, J. L. Vella, F. (2006). "Oxford Dictionary of Biochemistry and Molecular Biology (2nd Edition) – neomycin." Oxford University Press. (2006): 453. Knovel.com. Web. 18 Nov. 2014.
- ↑ Tsuji, K; Robertson, J. H.; Baas, R; McInnis, D. J. (1969). "Comparative study of responses to neomycins B and C by microbiological and gas-liquid chromatographic assay methods". Applied microbiology. 18 (3): 396–8. PMC 377991
 . PMID 4907002.
. PMID 4907002. - ↑ Zug, K. A.; Warshaw, E. M.; Fowler Jr, J. F.; Maibach, H. I.; Belsito, D. L.; Pratt, M. D.; Sasseville, D; Storrs, F. J.; Taylor, J. S.; Mathias, C. G.; Deleo, V. A.; Rietschel, R. L.; Marks, J (2009). "Patch-test results of the North American Contact Dermatitis Group 2005-2006". Dermatitis. 20 (3): 149–60. PMID 19470301.
- ↑ "The Nobel Prize in Physiology or Medicine 1952". Nobel Foundation. Retrieved 2008-10-29.
- ↑ "Neomycin." Pharmaceutical Manufacturing Encyclopedia (3rd edition) Volume 3. (2007): 2415–2416. Knovel.com. Web. 18 Nov. 2014.
- ↑ Jin, Yi; Watkins, Derrick; Degtyareva, Natalya N.; Green, Keith D.; Spano, Meredith N.; Garneau-Tsodikova, Sylvie; Arya, Dev P. (2016). "Arginine-Linked Neomycin B Dimers: Synthesis, rRNA Binding, and Resistance Enzyme Activity". MedChemComm. 7 (1): 164-169. doi:10.1039/C5MD00427F.
- ↑ Kaul, Malvika; Pilch, Daniel S. (2002). "Thermodynamics of Aminoglycoside−rRNA Recognition: The Binding of Neomycin-Class Aminoglycosides to the A Site of 16S rRNA". Biochemistry. 41 (24): 7695–706. PMID 12056901. doi:10.1021/bi020130f.
- ↑ Arya, Dev P; Coffee, R.Lane (2000). "DNA Triple Helix Stabilization by Aminoglycoside Antibiotics". Bioorganic & Medicinal Chemistry Letters. 10 (17): 1897–9. PMID 10987412. doi:10.1016/S0960-894X(00)00372-3.
- ↑ Arya, Dev P.; Coffee, R. Lane; Charles, I. (2001). "Neomycin-Induced Hybrid Triplex Formation". Journal of the American Chemical Society. 123 (44): 11093–4. PMID 11686727. doi:10.1021/ja016481j.