Flagellum
| Flagellum | |
|---|---|
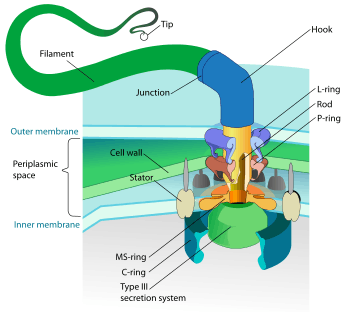 Structure of bacterial flagellum. | |
.jpg) SEM image of flagellated Chlamydomonas sp. (10000×) | |
| Identifiers | |
| TH | H1.00.01.1.01032 |
| FMA | 67472 |
A flagellum (/fləˈdʒɛləm/; plural: flagella) is a lash-like appendage that protrudes from the cell body of certain prokaryotic and eukaryotic cells. The word flagellum in Latin means whip. The primary role of the flagellum is locomotion, but it also often has function as a sensory organelle, being sensitive to chemicals and temperatures outside the cell.[1][2][3][4] Flagella are organelles defined by function rather than structure. Large differences occur between different types of flagella; the prokaryotic and eukaryotic flagella differ greatly in protein composition, structure, and mechanism of propulsion. However, both can be used for swimming.
An example of a flagellated bacterium is the ulcer-causing Helicobacter pylori, which uses multiple flagella to propel itself through the mucus lining to reach the stomach epithelium.[5] An example of a eukaryotic flagellate cell is the mammalian sperm cell, which uses its flagellum to propel itself through the female reproductive tract.[6] Eukaryotic flagella are structurally identical to eukaryotic cilia, although distinctions are sometimes made according to function and/or length.[7] Fimbriae and pili are also thin appendages, but have different functions and are usually smaller.
Types

Three types of flagella have so far been distinguished: bacterial, archaeal, and eukaryotic.
The main differences among these three types are:
- Bacterial flagella are helical filaments, each with a rotary motor at its base which can turn clockwise or counterclockwise.[8][9][10] They provide two of several kinds of bacterial motility.[11][12]
- Archaeal flagella (archaella) are superficially similar to bacterial flagella, but are different in many details and considered non-homologous.[13][14][15]
- Eukaryotic flagella—those of animal, plant, and protist cells—are complex cellular projections that lash back and forth. Eukaryotic flagella are classed along with eukaryotic motile cilia as undulipodia[16] to emphasize their distinctive wavy appendage role in cellular function or motility. Primary cilia are immotile, and are not undulipodia; they have a structurally different 9+0 axoneme rather than the 9+2 axoneme found in both flagella and motile cilia undulipodia.
Bacterial

Structure and composition
The bacterial flagellum is made up of the protein flagellin. Its shape is a 20-nanometer-thick hollow tube. It is helical and has a sharp bend just outside the outer membrane; this "hook" allows the axis of the helix to point directly away from the cell. A shaft runs between the hook and the basal body, passing through protein rings in the cell's membrane that act as bearings. Gram-positive organisms have two of these basal body rings, one in the peptidoglycan layer and one in the plasma membrane. Gram-negative organisms have four such rings: the L ring associates with the lipopolysaccharides, the P ring associates with peptidoglycan layer, the M ring is embedded in the plasma membrane, and the S ring is directly attached to the plasma membrane. The filament ends with a capping protein.[17][18]
The flagellar filament is the long, helical screw that propels the bacterium when rotated by the motor, through the hook. In most bacteria that have been studied, including the Gram-negative Escherichia coli, Salmonella typhimurium, Caulobacter crescentus, and Vibrio alginolyticus, the filament is made up of 11 protofilaments approximately parallel to the filament axis. Each protofilament is a series of tandem protein chains. However, Campylobacter jejuni has seven protofilaments.[19]
The basal body has several traits in common with some types of secretory pores, such as the hollow, rod-like "plug" in their centers extending out through the plasma membrane. Given the structural similarities between bacterial flagella and bacterial secretory systems, bacterial flagella may have evolved from the type-three secretion system; however, whether these pores are derived from the bacterial flagella or the bacterial secretory system is not known for certain.
Motor
The bacterial flagellum is driven by a rotary engine (Mot complex) made up of protein, located at the flagellum's anchor point on the inner cell membrane. The engine is powered by proton motive force, i.e., by the flow of protons (hydrogen ions) across the bacterial cell membrane due to a concentration gradient set up by the cell's metabolism (Vibrio species have two kinds of flagella, lateral and polar, and some are driven by a sodium ion pump rather than a proton pump[20]). The rotor transports protons across the membrane, and is turned in the process. The rotor alone can operate at 6,000 to 17,000 rpm, but with the flagellar filament attached usually only reaches 200 to 1000 rpm. The direction of rotation can be switched almost instantaneously, caused by a slight change in the position of a protein, FliG, in the rotor.[21] The flagellum is highly energy efficient and uses very little energy.[22] The exact mechanism for torque generation is still poorly understood.[23]Because the flagellar motor has no on-off switch, the protein epsE is used as a mechanical clutch to disengage the motor from the rotor, thus stopping the flagellum and allowing the bacterium to remain in one place.[24]
The cylindrical shape of flagella is suited to locomotion of microscopic organisms; these organisms operate at a low Reynolds number, where the viscosity of the surrounding water is much more important than its mass or inertia.[25]
The rotational speed of flagella varies in response to the intensity of the proton motive force, thereby permitting certain forms of speed control, and also permitting some types of bacteria to attain remarkable speeds in proportion to their size; some achieve roughly 60 cell lengths per second. At such a speed, a bacterium would take about 245 days to cover 1 km; although that may seem slow, the perspective changes when the concept of scale is introduced. In comparison to macroscopic life forms, it is very fast indeed when expressed in terms of number of body lengths per second. A cheetah, for example, only achieves about 25 body lengths per second.[26]
Through use of their flagella, E. coli is able to move rapidly towards attractants and away from repellents, by means of a biased random walk, with 'runs' and 'tumbles' brought about by rotating its flagellum counterclockwise and clockwise, respectively. The two directions of rotation are not identical (with respect to flagellum movement) and are selected by a molecular switch.[27]
Assembly
During flagellar assembly, components of the flagellum pass through the hollow cores of the basal body and the nascent filament. During assembly, protein components are added at the flagellar tip rather than at the base.[28] In vitro, flagellar filaments assemble spontaneously in a solution containing purified flagellin as the sole protein.[29]
Evolution
At least 10 protein components of the bacterial flagellum share homologous proteins with the type-three secretion system (TTSS),[30] hence one likely evolved from the other. Because the TTSS has a similar number of components as a flagellar apparatus (about 25 proteins), which one evolved first is difficult to determine. However, the flagellar system appears to involve more proteins overall, including various regulators and chaperones, hence it has been argued that flagella evolved from a TTSS. However, it has also been suggested[31] that the flagellum may have evolved first or the two structures evolved in parallel. Early single-cell organisms' need for motility (mobility) support that the more mobile flagella would be selected by evolution first,[31] but the TTSS evolving from the flagellum can be seen as 'reductive evolution', and receives no topological support from the phylogenetic trees.[32] The hypothesis that the two structures evolved separately from a common ancestor accounts for the protein similarities between the two structures, as well as their functional diversity.[33]
Flagella and the intelligent design debate
Some authors have argued that flagella cannot have evolved because they can only function properly when all proteins are in place.[34] In other words, the flagellar apparatus is "irreducibly complex". This has long been debunked, because many proteins can be deleted or mutated and the flagellum still works, though sometimes at reduced efficiency.[35] In addition, the composition of flagella is surprisingly diverse across bacteria, with many proteins only found in some species, but not others.[36] Hence, the flagellar apparatus is clearly very flexible in evolutionary terms and perfectly able to lose or gain protein components. For instance, a number of mutations have been found that increase the motility of E. coli.[37]. Additional evidence for the evolution of bacterial flagella includes the existence of vestigial flagella, intermediate forms of flagella and patterns of similarities among flagellar protein sequences, including the observation that almost all of the core flagellar proteins have known homologies with non-flagellar proteins.[38] Furthermore, several processes have been identified as playing important roles in flagellar evolution, including self-assembly of simple repeating subunits, gene duplication with subsequent divergence, recruitment of elements from other systems (‘molecular bricolage’) and recombination. [39]
Flagellar arrangement schemes

Different species of bacteria have different numbers and arrangements of flagella.
- Monotrichous bacteria have a single flagellum (e.g., Vibrio cholerae).
- Lophotrichous bacteria have multiple flagella located at the same spot on the bacterial surfaces which act in concert to drive the bacteria in a single direction. In many cases, the bases of multiple flagella are surrounded by a specialized region of the cell membrane, the so-called polar organelle.
- Amphitrichous bacteria have a single flagellum on each of two opposite ends (only one flagellum operates at a time, allowing the bacterium to reverse course rapidly by switching which flagellum is active).
- Peritrichous bacteria have flagella projecting in all directions (e.g., E. coli).
In certain large forms of Selenomonas, more than 30 individual flagella are organized outside the cell body, helically twining about each other to form a thick structure (easily visible with the light microscope) called a "fascicle".
Other bacteria, such as most spirochetes, have two or more specialized flagella (endoflagella) arising from opposite poles of the cell, which together constitute the so-called "axial filament" that is located within the periplasmic space between the flexible cell wall and an outer sheath. The rotation of the axial filament relative to the cell body causes the entire bacterium to move forward in a corkscrew-like motion, even through material viscous enough to prevent the passage of normally flagellated bacteria.
Counterclockwise rotation of a monotrichous polar flagellum pushes the cell forward with the flagellum trailing behind, much like a corkscrew moving inside cork. Indeed, water on the microscopic scale is highly viscous, very different from our daily experience of water.
Flagella are left-handed helices, and bundle and rotate together only when rotating counterclockwise. When some of the rotors reverse direction, the flagella unwind and the cell starts "tumbling". Even if all flagella would rotate clockwise, they likely will not form a bundle, due to geometrical, as well as hydrodynamic reasons.[40][41] Such "tumbling" may happen occasionally, leading to the cell seemingly thrashing about in place, resulting in the reorientation of the cell. The clockwise rotation of a flagellum is suppressed by chemical compounds favorable to the cell (e.g. food), but the motor is highly adaptive to this. Therefore, when moving in a favorable direction, the concentration of the chemical attractant increases and "tumbles" are continually suppressed; however, when the cell's direction of motion is unfavorable (e.g., away from a chemical attractant), tumbles are no longer suppressed and occur much more often, with the chance that the cell will be thus reoriented in the correct direction.
In some Vibrio spp. (particularly Vibrio parahemolyticus[42]) and related proteobacteria such as Aeromonas, two flagellar systems co-exist, using different sets of genes and different ion gradients for energy. The polar flagella are constitutively expressed and provide motility in bulk fluid, while the lateral flagella are expressed when the polar flagella meet too much resistance to turn.[43][44][45][46][47][48] These provide swarming motility on surfaces or in viscous fluids.
Archaeal
The archaellum possessed by some members of domain Archea is superficially similar to the bacterial flagellum; in the 1980s, they were thought to be homologous on the basis of gross morphology and behavior.[49] Both flagella and archaella consist of filaments extending outside the cell, and rotate to propel the cell. Archaeal flagella have a unique structure which lacks a central channel. Similar to bacterial type IV pilins, the archaeal flagellins (archaellins) are made with class 3 signal peptides and they are processed by a type IV prepilin peptidase-like enzyme. The archaellins are typically modified by the addition of N-linked glycans which are necessary for proper assembly and/or function.[4]
Discoveries in the 1990s revealed numerous detailed differences between the archaeal and bacterial flagella. These include:
- Bacterial flagella are motorized by a flow of H+ ions (or occasionally Na+ ions); archaeal flagella are almost certainly powered by ATP. The torque-generating motor that powers rotation of the archaeal flagellum has not been identified.
- While bacterial cells often have many flagellar filaments, each of which rotates independently, the archaeal flagellum is composed of a bundle of many filaments that rotates as a single assembly.
- Bacterial flagella grow by the addition of flagellin subunits at the tip; archaeal flagella grow by the addition of subunits to the base.
- Bacterial flagella are thicker than archaella, and the bacterial filament has a large enough hollow "tube" inside that the flagellin subunits can flow up the inside of the filament and get added at the tip; the archaellum is too thin (12-15 nm) to allow this.[50]
- Many components of bacterial flagella share sequence similarity to components of the type III secretion systems, but the components of bacterial flagella and archaella share no sequence similarity. Instead, some components of archaella share sequence and morphological similarity with components of type IV pili, which are assembled through the action of type II secretion systems (the nomenclature of pili and protein secretion systems is not consistent).[50]
These differences could mean that the bacterial flagella and archaella could be a classic case of biological analogy, or convergent evolution, rather than homology. However, in comparison to the decades of well-publicized study of bacterial flagella (e.g. by Howard Berg),[51] archaella have only recently begun to garner scientific attention.
Eukaryotic




Terminology
Aiming to emphasize the distinction between the bacterial flagella and the eukaryotic cilia and flagella, some authors attempted to replace the name of these two eukaryotic structures with "undulipodia" (e.g., all papers by Margulis since the 1970s)[52] or "cilia" for both (e.g., Hülsmann, 1992;[53] Adl et al., 2012;[54] most papers of Cavalier-Smith), preserving "flagella" for the bacterial structure. However, the discriminative usage of the terms "cilia" and "flagella" for eukaryotes adopted in this article is still common (e.g., Andersen et al., 1991;[55] Leadbeater et al., 2000).[56]
Internal structure
A eukaryotic flagellum is a bundle of nine fused pairs of microtubule doublets surrounding two central single microtubules. The so-called "9 + 2" structure is characteristic of the core of the eukaryotic flagellum called an axoneme. At the base of a eukaryotic flagellum is a basal body, "blepharoplast" or kinetosome, which is the microtubule organizing center for flagellar microtubules and is about 500 nanometers long. Basal bodies are structurally identical to centrioles. The flagellum is encased within the cell's plasma membrane, so that the interior of the flagellum is accessible to the cell's cytoplasm.
Besides the axoneme and basal body, relatively constant in morphology, other internal structures of the flagellar apparatus are the transition zone (where the axoneme and basal body meet) and the root system (microtubular or fibrilar structures which extends from the basal bodies into the cytoplasm), more variable and useful as indicators of phylogenetic relationships of eukaryotes. Other structures, more uncommon, are the paraflagellar (or paraxial, paraxonemal) rod, the R fiber, and the S fiber.[57] For surface structures, see below.
Mechanism
Each of the outer 9 doublet microtubules extends a pair of dynein arms (an "inner" and an "outer" arm) to the adjacent microtubule; these produce force through ATP hydrolysis. The flagellar axoneme also contains radial spokes, polypeptide complexes extending from each of the outer nine microtubule doublets towards the central pair, with the "head" of the spoke facing inwards. The radial spoke is thought to be involved in the regulation of flagellar motion, although its exact function and method of action are not yet understood.
Flagella vs cilia

The regular beat patterns of eukaryotic cilia and flagella generate motion on a cellular level. Examples range from the propulsion of single cells such as the swimming of spermatozoa to the transport of fluid along a stationary layer of cells such as in the respiratory tract. Though eukaryotic flagella and motile cilia are ultrastructurally identical, the beating pattern of the two organelles can be different. In the case of flagella, the motion is often planar and wave-like, whereas the motile cilia often perform a more complicated three-dimensional motion with a power and recovery stroke.
Intraflagellar transport
Intraflagellar transport, the process by which axonemal subunits, transmembrane receptors, and other proteins are moved up and down the length of the flagellum, is essential for proper functioning of the flagellum, in both motility and signal transduction.[58]
Evolution and occurrence
Eukaryotic flagella or cilia, probably an ancestral characteristic,[59] are widespread in almost all groups of eukaryotes, as a relatively perennial condition, or as a flagellated life cycle stage (e.g., zoids, gametes, zoospores, which may be produced continually or not).[60][61][62]
The first situation is found either in specialized cells of multicellular organisms (e.g., the choanocytes of sponges, or the ciliated epithelia of metazoans), as in ciliates and many eukaryotes with a "flagellate condition" (or "monadoid level of organization", see Flagellata, an artificial group).
Flagellated lifecycle stages are found in many groups, e.g., many green algae (zoospores and male gametes), bryophytes (male gametes), pteridophytes (male gametes), some gymnosperms (cycads and Ginkgo, as male gametes), centric diatoms (male gametes), brown algae (zoospores and gametes), oomycetes (assexual zoospores and gametes), hyphochytrids (zoospores), labyrinthulomycetes (zoospores), some apicomplexans (gametes), some radiolarians (probably gametes),[63] foraminiferans (gametes), plasmodiophoromycetes (zoospores and gametes), myxogastrids (zoospores), metazoans (male gametes), and chytrid fungi (zoospores and gametes).
Flagella or cilia are completely absent in some groups, probably due to a loss rather than being a primitive condition. The loss of cilia occurred in red algae, some green algae (Zygnematophyceae), the gymnosperms except cycads and Ginkgo, angiosperms, pennate diatoms, some apicomplexans, some amoebozoans, in the sperm of some metazoans,[64] and in fungi (except chytrids).
Typology
A number of terms related to flagella or cilia are used to characterize eukaryotes.[65][66][67][68][69] According to surface structures present, flagella may be:
- whiplash flagella (= smooth, acronematic flagella): without hairs, e.g., in Opisthokonta
- hairy flagella (= tinsel, flimmer, pleuronematic flagella): with hairs (= mastigonemes sensu lato), divided in:
- with fine hairs (= non tubular, or simple hairs): occurs in Euglenophyceae, Dinoflagellata, some Haptophyceae (Pavlovales)
- with stiff hairs (= tubular hairs, retronemes, mastigonemes sensu stricto), divided in:
- bipartite hairs: with two regions. Occurs in Cryptophyceae, Prasinophyceae, and some Heterokonta
- tripartite (= straminipilous) hairs: with three regions (a base, a tubular shaft, and one or more terminal hairs). Occurs in most Heterokonta
- stichonematic flagella: with a single row of hairs
- pantonematic flagella: with two rows of hairs
- acronematic: flagella with a single, terminal mastigoneme or flagellar hair (e.g., bodonids);[70] some authors use the term as synonym of whiplash
- with scales: e.g., Prasinophyceae
- with spines: e.g., some brown algae
- with undulating membrane: e.g., some kinetoplastids, some parabasalids
- with proboscis (trunk-like protrusion of the cell): e.g., apusomonads, some bodonids[71]
According to the number of flagella, cells may be (remembering that some authors use "ciliated" instead of "flagellated":[62][72]
- uniflagellated: e.g., most Opisthokonta
- biflagellated: e.g., all Dinoflagellata, the gametes of Charophyceae, of most bryophytes and of some metazoans[73]
- triflagellated: e.g., the gametes of some Foraminifera
- quadriflagellated: e.g., some Prasinophyceae, Collodictyonidae
- octoflagellated: e.g., some Diplomonada, some Prasinophyceae
- multiflagellated: e.g., Opalinata, Ciliophora, Stephanopogon, Parabasalida, Hemimastigophora, Caryoblastea, Multicilia, the gametes (or zoids) of Oedogoniales (Chlorophyta), some pteridophytes and some gymnosperms
According to the place of insertion of the flagella:[74]
- opisthokont: cells with flagella inserted posteriorlly, e.g., in Opisthokonta (Vischer, 1945). In Haptophyceae, flagella are laterally to terminally inserted, but are directed posteriorly during rapid swimming.[75]
- akrokont: cells with flagella inserted apically
- subakrokont: cells with flagella inserted subapically
- pleurokont: cells with flagella inserted laterally
According to the beating pattern:
- gliding: a flagellum that trails on the substrate[71]
- heterodynamic: flagella with different beating patterns (usually with one flagellum functioning in food capture and the other functioning in gliding, anchorage, propulsion or “steering”)[76]
- isodynamic: flagella beating with the same patterns
Other terms related to the flagellar type:
- isokont: cells with flagella of equal length. It was also formerly used to refer to the Chlorophyta
- anisokont: cells with flagella of unequal length, e.g., some Euglenophyceae and Prasinophyceae
- heterokont: term introduced by Luther (1899) to refer to the Xanthophyceae, due to the pair of flagella of unequal length. It has taken on a specific meaning in referring to cells with an anterior straminipilous flagellum (with tripartite mastigonemes, in one or two rows) and a posterior usually smooth flagellum. It is also used to refer to the taxon Heterokonta
- stephanokont: cells with a crown of flagella near its anterior end, e.g., the gametes and spores of Oedogoniales, the spores of some Bryopsidales. Term introduced by Blackman & Tansley (1902) to refer to the Oedogoniales
- akont: cells without flagella. It was also used to refer to taxonomic groups, as Aconta or Akonta: the Zygnematophyceae and Bacillariophyceae (Oltmanns, 1904), or the Rhodophyceae (Christensen, 1962)
See also
- Archaellum
- Cilium
- Evolution of flagella
- Ciliopathy
- Rotating locomotion in living systems
- Undulipodium
References
- ↑ Wang, Qingfeng; Suzuki, Asaka; Mariconda, Susana; Porwollik, Steffen; Harshey, Rasika M (2005). "Sensing wetness: A new role for the bacterial flagellum". The EMBO Journal. 24 (11): 2034–42. PMC 1142604
 . PMID 15889148. doi:10.1038/sj.emboj.7600668.
. PMID 15889148. doi:10.1038/sj.emboj.7600668. - ↑ Bardy SL, Ng SY, Jarrell KF (February 2003). "Prokaryotic motility structures". Microbiology (Reading, Engl.). 149 (Pt 2): 295–304. PMID 12624192. doi:10.1099/mic.0.25948-0.
- ↑ Lefebvre PA; Lefebvre, PA (2001). "Assembly and Motility of Eukaryotic Cilia and Flagella. Lessons from Chlamydomonas reinhardtii". Plant Physiol. 127 (4): 1500–1507. PMC 1540183
 . PMID 11743094. doi:10.1104/pp.010807.
. PMID 11743094. doi:10.1104/pp.010807. - 1 2 Jarrell, K (editor) (2009). Pili and Flagella: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-48-6.
- ↑ Lacy BE, Rosemore J (October 2001). "Helicobacter pylori: ulcers and more: the beginning of an era" (abstract page). J. Nutr. 131 (10): 2789S–2793S. PMID 11584108.
- ↑ Malo AF, Gomendio M, Garde J, Lang-Lenton B, Soler AJ, Roldan ER (June 2006). "Sperm design and sperm function". Biol. Lett. 2 (2): 246–9. PMC 1618917
 . PMID 17148374. doi:10.1098/rsbl.2006.0449.
. PMID 17148374. doi:10.1098/rsbl.2006.0449. - ↑ Haimo LT, Rosenbaum JL (December 1981). "Cilia, flagella, and microtubules". J. Cell Biol. 91 (3 Pt 2): 125s–130s. PMC 2112827
 . PMID 6459327. doi:10.1083/jcb.91.3.125s.
. PMID 6459327. doi:10.1083/jcb.91.3.125s. - ↑ Silverman M, Simon M (1974). "Flagellar rotation and the mechanism of bacterial motility". Nature. 249 (452): 73–74. PMID 4598030. doi:10.1038/249073a0.
- ↑ Meister GL, Berg HC (1987). "Rapid rotation of flagellar bundles in swimming bacteria". Nature. 325 (6105): 637–640. doi:10.1038/325637a0.
- ↑ Berg HC, Anderson RA (1973). "Bacteria Swim by Rotating their Flagellar Filaments". Nature. 245 (5425): 380–382. PMID 4593496. doi:10.1038/245380a0.
- ↑ Jahn TL, Bovee EC (1965). "Movement and Locomotion of Microorganisms". Annual Review of Microbiology. 19: 21–58. PMID 5318439. doi:10.1146/annurev.mi.19.100165.000321.
- ↑ Harshey RM (2003). "Bacterial Motility on a Surface: Many Ways to a Common Goal". Annual Review of Microbiology. 57: 249–273. PMID 14527279. doi:10.1146/annurev.micro.57.030502.091014.
- ↑ Ng SY, Chaban B, Jarrell KF (2006). "Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications". J. Mol. Microbiol. Biotechnol. 11 (3–5): 167–91. PMID 16983194. doi:10.1159/000094053.
- ↑ Metlina AL (2004). "Bacterial and archaeal flagella as prokaryotic motility organelles". Biochemistry Mosc. 69 (11): 1203–12. PMID 15627373. doi:10.1007/s10541-005-0065-8.
- ↑ Jarrell (2009). "Archaeal Flagella and Pili". Pili and Flagella: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-48-6.
- ↑ A Dictionary of Biology, 2004, accessed 2011-01-01.
- ↑ Macnab RM (2003). "How bacteria assemble flagella". Annu. Rev. Microbiol. 57: 77–100. PMID 12730325. doi:10.1146/annurev.micro.57.030502.090832.
- ↑ Diószeghy Z, Závodszky P, Namba K, Vonderviszt F (2004). "Stabilization of flagellar filaments by HAP2 capping". FEBS Lett. 568 (1–3): 105–9. PMID 15196929. doi:10.1016/j.febslet.2004.05.029.
- ↑ Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH (2008). "Divergence of quaternary structures among bacterial flagellar filaments". Science. 320 (5874): 382–5. PMID 18420936. doi:10.1126/science.1155307.
- ↑ Atsumi T, McCarter L, Imae Y (1992). "Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces". Nature. 355 (6356): 182–4. PMID 1309599. doi:10.1038/355182a0.
- ↑ Dean, Tim. "Inside nature’s most efficient motor: the flagellar", Australian Life Scientist, 2 August 2010. Retrieved on 2013-08-28.
- ↑ Unlocking the secrets of nature's nanomotor Nikkei Asian Review, 2014.
- ↑ Thierry, Mora; Howard, Yu. "Steps in the Bacterial Flagellar Motor". PLOS. Retrieved 17 May 2017.
- ↑ Whitfield, John (19 June 2008). "Bacterial engines have their own clutch". Nature News. doi:10.1038/news.2008.903. Retrieved 17 May 2017.
- ↑ Dusenbery DB (2009). "Chapter 13". Living at Micro Scale: The Unexpected Physics of Being Small. Cambridge: Harvard University Press. ISBN 0-674-03116-4.
- ↑ Hildebrand, Milton (November 1959). "Motions of the running Cheetah and Horse". Journal of Mammalogy. 44 (4): 481–495. JSTOR 1376265. Although according to Cheetah, Luke Hunter and Dave Hamman, (Struik Publishers, 2003), pp. 37–38, the cheetah's fastest recorded speed was 110 km/h (68 mph).
- ↑ Meadows, Robin (10 May 2011). "How Bacteria Shift Gears". PLoS Biology. doi:10.1371/journal.pbio.1001061.
- ↑ Minamino T, Imada K, Namba K (2008). "Mechanisms of type III protein export for bacterial flagellar assembly". Mol. Biosyst. 4 (11): 1105–15. PMID 18931786. doi:10.1039/b808065h.
- ↑ Asakura S, Eguchi G, Iino T (1964). "Reconstitution of Bacterial Flagella in Vitro". J. Mol. Biol. 10: 42–56. PMID 14222895. doi:10.1016/S0022-2836(64)80026-7.
- ↑ Pallen MJ, Matzke NJ (2006). "From The Origin of Species to the origin of bacterial flagella". Nat Rev Microbiol. 4 (10): 784–90. PMID 16953248. doi:10.1038/nrmicro1493.
- 1 2 Saier, M (2004). "Evolution of bacterial type III protein secretion systems". Trends in Microbiology. 12 (3): 113–115. PMID 15001186. doi:10.1016/j.tim.2004.01.003.
- ↑ Gophna U, Ron EZ, Graur D (July 2003). "Bacterial type III secretion systems are ancient and evolved by multiple horizontal-transfer events". Gene. 312: 151–63. PMID 12909351. doi:10.1016/S0378-1119(03)00612-7.
- ↑ Guttman D, McCann H (2008). "Evolution of the type III secretion system and its effectors in plant-microbe interactions". New Phytologist. 177 (1): 33–47. doi:10.1111/J.1469-8137.2007.02293.X.
- ↑ Behe, M. (2007) The Edge of Evolution. Free Press, New York
- ↑ Rajagopala SV, Titz B, Goll J, Parrish JR, Wohlbold K, McKevitt MT, Palzkill T, Mori H, Finley RL Jr, Uetz P (2007). "The protein network of bacterial motility". Mol Syst Biol. 3: 128. PMC 1943423
 . PMID 17667950. doi:10.1038/msb4100166.
. PMID 17667950. doi:10.1038/msb4100166. - ↑ Titz B, Rajagopala SV, Ester C, Häuser R, Uetz P (Nov 2006). "Novel conserved assembly factor of the bacterial flagellum". J Bacteriol. 188 (21): 7700–6. PMC 1636259
 . PMID 16936039. doi:10.1128/JB.00820-06.
. PMID 16936039. doi:10.1128/JB.00820-06. - ↑ Kakkanat, Asha; Phan, Minh-Duy; Lo, Alvin W.; Beatson, Scott A.; Schembri, Mark A. (2017-05-10). "Novel genes associated with enhanced motility of Escherichia coli ST131". PLOS ONE. 12 (5): e0176290. ISSN 1932-6203. doi:10.1371/journal.pone.0176290.
- ↑ Pallen MJ, Matzke NJ (2006). "From The Origin of Species to the origin of bacterial flagella". Nature Reviews Microbiology. 4: 784. PMID 16953248. doi:10.1038/nrmicro1493.
- ↑ Pallen MJ, Gophna U (2007). "Bacterial Flagella and Type III Secretion: Case Studies in the Evolution of Complexity". Gene and Protein Evolution. 3: 30. PMID 18753783. doi:10.1159/000107602.
- ↑ Kim M, Bird JC, Van Parys AJ, Breuer KS, Powers TR (December 2003). "A macroscopic scale model of bacterial flagellar bundling". Proc. Natl. Acad. Sci. U.S.A. 100 (26): 15481–5. PMC 307593
 . PMID 14671319. doi:10.1073/pnas.2633596100.
. PMID 14671319. doi:10.1073/pnas.2633596100. - ↑ Macnab RM (January 1977). "Bacterial flagella rotating in bundles: a study in helical geometry". Proc. Natl. Acad. Sci. U.S.A. 74 (1): 221–5. PMC 393230
 . PMID 264676. doi:10.1073/pnas.74.1.221.
. PMID 264676. doi:10.1073/pnas.74.1.221. - ↑ Kim YK, McCarter LL (2000). "Analysis of the Polar Flagellar Gene System of Vibrio parahaemolyticus". Journal of Bacteriology. 182 (13): 3693–3704. PMC 94540
 . PMID 10850984. doi:10.1128/JB.182.13.3693-3704.2000.
. PMID 10850984. doi:10.1128/JB.182.13.3693-3704.2000. - ↑ Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, Homma M (1 August 1996). "Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus". Journal of Bacteriology. 178 (16): 5024–5026. PMC 178290
 . PMID 8759871.
. PMID 8759871. - ↑ McCarter LL (2004). "Dual Flagellar Systems Enable Motility under Different Circumstances". Journal of Molecular Microbiology and Biotechnology. 7 (1–2): 18–29. PMID 15170400. doi:10.1159/000077866.
- ↑ Merino S, Shaw JG, Tomás JM (2006). "Bacterial lateral flagella: an inducible flagella system". FEMS Microbiol Lett. 263 (2): 127–35. PMID 16978346. doi:10.1111/j.1574-6968.2006.00403.x.
- ↑ Belas R, Simon M, Silverman M (1986). "Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus". J Bacteriol. 167 (1): 210–8. PMC 212863
 . PMID 3013835.
. PMID 3013835. - ↑ Canals R, Altarriba M, Vilches S, Horsburgh G, Shaw JG, Tomás JM, Merino S (2006). "Analysis of the Lateral Flagellar Gene System of Aeromonas hydrophila AH-3". Journal of Bacteriology. 188 (3): 852–862. PMC 1347325
 . PMID 16428388. doi:10.1128/JB.188.3.852-862.2006.
. PMID 16428388. doi:10.1128/JB.188.3.852-862.2006. - ↑ Canals R, Ramirez S, Vilches S, Horsburgh G, Shaw JG, Tomás JM, Merino S (January 2006). "Polar Flagellum Biogenesis in Aeromonas hydrophila". J. Bacteriol. 188 (2): 542–55. PMC 1347287
 . PMID 16385045. doi:10.1128/JB.188.2.542-555.2006.
. PMID 16385045. doi:10.1128/JB.188.2.542-555.2006. - ↑ Cavalier-Smith T (1987). "The origin of eukaryotic and archaebacterial cells". Ann. N. Y. Acad. Sci. 503 (1): 17–54. PMID 3113314. doi:10.1111/j.1749-6632.1987.tb40596.x.
- 1 2 Ghosh A, Albers SV (January 2011). "Assembly and function of the archaeal flagellum". Biochem. Soc. Trans. 39 (1): 64–9. PMID 21265748. doi:10.1042/BST0390064.
- ↑ Berg, Howard C. (2003). E. coli in motion (1. Aufl. ed.). New York: Springer. ISBN 9780387008882.
- ↑ Taylor, F. J. [The collapse of the two-kingdom system, the rise of protistology and the founding of the International Society for Evolutionary Protistology (ISEP)]. International journal of systematic and evolutionary microbiology 53: 1707-1714.
- ↑ Hülsmann N (1992). "Undulipodium: end of a useless discussion". Eur J Protistol. 28: 253–257. PMID 23195228. doi:10.1016/s0932-4739(11)80231-2.
- ↑ Adl SM, et al. (2012). "The revised classification of eukaryotes" (PDF). Journal of Eukaryotic Microbiology. 59 (5): 429–514. Archived from the original (PDF) on 16 December 2013.
- ↑ Andersen RA, Barr DJ, Lynn DH, Melkonian M, Moestrup Ø, Sleigh MA (1991). "Terminology and nomenclature of the cytoskeletal elements associated with the flagellar/ciliary apparatus in protists". Protoplasma. 164 (1–3): 1–8. doi:10.1007/bf01320809.
- ↑ Leadbeater, B.S.C., Green, J.C. (eds.). The Flagellates. Unity, diversity, and evolution. London: Taylor and Francis.
- ↑ Barsanti, L. & Gualtieri, P. (2006). pp. 63-84.
- ↑ Pazour GJ (October 2004). "Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease". J. Am. Soc. Nephrol. 15 (10): 2528–36. PMID 15466257. doi:10.1097/01.ASN.0000141055.57643.E0.
- ↑ Yubuki N, Leander BS (2013). "Evolution of microtubule organizing centers across the tree of eukaryotes". The Plant Journal. 75 (2): 230–244. doi:10.1111/tpj.12145.
- ↑ Raven, J.A. 2000. The flagellate condition. In: (B.S.C. Leadbeater and J.C. Green, eds) The flagellates. Unity, diversity, and evolution. The Systematics Association Special Volume 59. Taylor and Francis, London. pp. 269–287.
- ↑ Webster, J & Weber, R (2007). Introduction to Fungi (3rd ed.). Cambridge: Cambridge University Press. pp. 23–24,
- 1 2 Adl et al. (2012).
- ↑ Lahr DJ, Parfrey LW, Mitchell EA, Katz LA, Lara E (July 2011). "The chastity of amoebae: re-evaluating evidence for sex in amoeboid organisms". Proc. Biol. Sci. 278: 2081–90. PMC 3107637
 . PMID 21429931. doi:10.1098/rspb.2011.0289.
. PMID 21429931. doi:10.1098/rspb.2011.0289. - ↑ Austin, C. R. (1995). Evolution of human gametes: spermatozoa. In: Grudzinskas, J. G., Yovich, J. L. (eds). Gametes: the spermatozoon. Cambridge University Press,
- ↑ Webster & Weber (2007).
- ↑ South, G.R. & Whittick, A. (1987). Introduction to Phycology. Blackwell Scientific Publications, Oxford. p. 65, .
- ↑ Barsanti, Laura; Gualtieri, Paolo (2006). Algae: anatomy, biochemistry, and biotechnology. Florida, USA: CRC Press. pp. 60-63,
- ↑ Dodge, J.D. (1973). The Fine Structure of Algal Cells. Academic Press, London. pp. 57-79,
- ↑ Lee, R. E. (2008). Phycology (4th ed.). Cambridge University Press. p. 7, .
- ↑ Corliss, J.O., Lom, J. (2002). An annotated glossary of protozoological terms. In: Lee J.J., Leedale G.F., Bradbury P. (eds.). An illustrated guide to the protozoa, second edition, pp. 1346–1385. Society of Protozoologists, Lawrence.
- 1 2 Jeuck, A., & Arndt, H. (2013). A short guide to common heterotrophic flagellates of freshwater habitats based on the morphology of living organisms. Protist, 164 (6): 842-860, .
- ↑ Sleigh, M. (1989). Protozoa and other Protists. Edward Arnold, London. pp 98-99, .
- ↑ Austin, C. R. (1995).
- ↑ Sparrow, F.K. (1960). Aquatic phycomycetes. 2nd ed. Ann Arbor, Michigan: University of Michigan Press, p. 15, .
- ↑ Hibberd DJ (1976). "The ultrastructure and taxonomy of the Chrysophyceae and Prymnesiophyceae (Haptophyceae): a survey with some new observations on the ultrastructure of the Chrysophyceae". Journal of the Linnean Society of London, Botany. 72: 55–80. doi:10.1111/j.1095-8339.1976.tb01352.x.
- ↑ Sleigh, M.A. (1985). Origin and evolution of flagellar movement. Cell Motil. 5:137-138, .
External links
| Wikimedia Commons has media related to Flagella. |
- "Molecular Machines Museum Index". Access Research Network. 2001. Retrieved 2008-05-18.
- Berg, Howard C. (January 2000). "Motile Behavior of Bacteria". Physics Today. 53 (1): 24–29. doi:10.1063/1.882934.
- Charles Lindemann (2008-04-04). "Mechanisms of sperm motility". Oakland University. Retrieved 2008-05-18.
- Purcell, E.M. (1977). "Life at Low Reynolds Number" (PDF). American Journal of Physics. 45 (1): 3–11. doi:10.1119/1.10903.
- N. J. Matzke (2003-11-10). "Evolution in (Brownian) space: a model for the origin of the bacterial flagellum". www.talkdesign.org.
- "What is flagella".
![]() This article incorporates text from a publication now in the public domain: Chambers, Ephraim, ed. (1728). "article name needed". Cyclopædia, or an Universal Dictionary of Arts and Sciences (first ed.). James and John Knapton, et al.
This article incorporates text from a publication now in the public domain: Chambers, Ephraim, ed. (1728). "article name needed". Cyclopædia, or an Universal Dictionary of Arts and Sciences (first ed.). James and John Knapton, et al.
