Alpha-enolase
Enolase 1 (ENO1), more commonly known as alpha-enolase, is a glycolytic enzyme expressed in most tissues, one of the isozymes of enolase. Each isoenzyme is a homodimer composed of 2 alpha, 2 gamma, or 2 beta subunits, and functions as a glycolytic enzyme. Alpha-enolase, in addition, functions as a structural lens protein (tau-crystallin) in the monomeric form. Alternative splicing of this gene results in a shorter isoform that has been shown to bind to the c-myc promoter and function as a tumor suppressor. Several pseudogenes have been identified, including one on the long arm of chromosome 1. Alpha-enolase has also been identified as an autoantigen in Hashimoto encephalopathy.[3]
Structure
ENO1 is one of three enolase isoforms, the other two being ENO2 (ENO-γ) and ENO3 (ENO-β).[4] Each isoform is a protein subunit that can hetero- or homodimerize to form αα, αβ, αγ, ββ, and γγ dimers.[5] The ENO1 gene spans 18 kb and lacks a TATA box while possessing multiple transcription start sites.[6] A hypoxia-responsive element can be found in the ENO1 promoter and allows the enzyme to function in aerobic glycolysis and contribute to the Warburg effect in tumor cells.[7]
Relationship to Myc-binding protein-1
The mRNA transcript of the ENO1 gene can be alternatively translated into a cytoplasmic protein, with a molecular weight of 48 kDa, or a nuclear protein, with a molecular weight of a 37 kDa.[7][8] The nuclear form was previously identified as Myc-binding protein-1 (MBP1), which downregulates the protein level of the c-myc protooncogene.[8][9] A start codon at codon 97 of ENO1 and a Kozak consensus sequence were found preceding the 3' region of ENO1 encoding the MBP1 protein. In addition, the N-terminal region of the MBP1 protein it critical to DNA binding and, thus, its inhibitory function.[8]
Function
As an enolase, ENO1 is a glycolytic enzyme the catalyzes the conversion of 2-phosphoglycerate to phosphoenolpyruvate.[4][7][10] This isozyme is ubiquitously expressed in adult human tissues, including liver, brain, kidney, and spleen.[4] Within cells, ENO1 predominantly localizes to the cytoplasm, though an alternatively translated form is localizes to the nucleus.[4][7] Its nuclear form, also known as MBP1, functions solely as a tumor suppressor by binding and inhibiting the c-myc protooncogene promoter, and lacks the glycolytic enzyme activity of the cytoplasmic form.[8] ENO1 also plays a role in other functions, including a cell surface receptor for plasminogen on pathogens, such as streptococci, and activated immune cells, leading to systemic infection or tissue invasion; an oxidative stress protein in endothelial cells; a lens crystalline; a heat shock protein; and a binding partner of cytoskeletal and chromatin structures to aid in transcription.[7][8][10][11][12]
Clinical significance
Cancer
ENO1 overexpression has been associated with multiple tumors, including glioma, neuroendocrine tumors, neuroblastoma, pancreatic cancer, prostate cancer, cholangiocarcinoma, thyroid carcinoma, lung cancer, hepatocellular carcinoma, and breast cancer.[4][7][12][13] In many of these tumors, ENO1 promoted cell proliferation by regulating the PI3K/AKT signaling pathway and induced tumorigenesis by activating plasminogen.[4][7] Moreover, ENO1 is expressed on the tumor cell surface during pathological conditions such as inflammation, autoimmunity, and malignancy. Its role as a plasminogen receptor leads to extracellular matrix degradation and cancer invasion.[7][12][13] Due to its surface expression, targeting surface ENO1 enables selective targeting of tumor cells while leaving the ENO1 inside normal cells functional.[7] Moreover, in tumors such as Non-Hodgkin's Lymphomas (NHLs) and breast cancer, inhibition of ENO1 expression decreased tolerance to hypoxia while increasing sensitivity to radiation therapy, thus indicating that ENO1 may have aided chemoresistance.[4][10] Considering these factors, ENO1 holds great potential to serve as an effective therapeutic target for treating many types of tumors in patients.[4][10][12]
ENO1 is located on the 1p36 tumor suppressor locus near MIR34A which is homozygously deleted in Glioblastoma, Hepatocellular carcinoma and Cholangiocarcinoma.[14][15][16] The co-deletion of ENO1 is a passenger event with the resultant tumor cells being entirely dependent on ENO2 for the execution of glycolysis.[17][18] Tumor cells with such deletions are exceptionally sensitive towards ablation of ENO2.[17][18] Inhibition of ENO2 in ENO1-homozygously deleted cancer cells constitutes an example of synthetic lethality treatment for cancer.
Autoimmune disease
ENO1 has been detected in serum drawn from children diagnosed with juvenile idiopathic arthritis.[19]
Alpha-enolase has been identified as an autoantigen in Hashimoto's encephalopathy.[20] Single studies have also identified it as an autoantigen associated with severe asthma[21] and a putative target antigen of anti-endothelial cell antibody in Behçet's disease.[22] Reduced expression of the enzyme has been found in the corneal epithelium of people suffering from keratoconus.[23][24]
Gastrointestinal disease
CagA protein was found to activate ENO1 expression through activating the Src and MEK/ERK pathways as a mechanism for H. pylori-mediated gastric diseases.[13]
Hemolytic anemia
Enolase deficiency is a rare inborn error of metabolism disease, leads to hemolytic anemia in affected homozygous carriers of loss of function mutations in ENO1.[25] As with other glycolysis enzyme deficiency diseases, the condition is aggravated by redox-cycling agents such as nitrofurantoin.
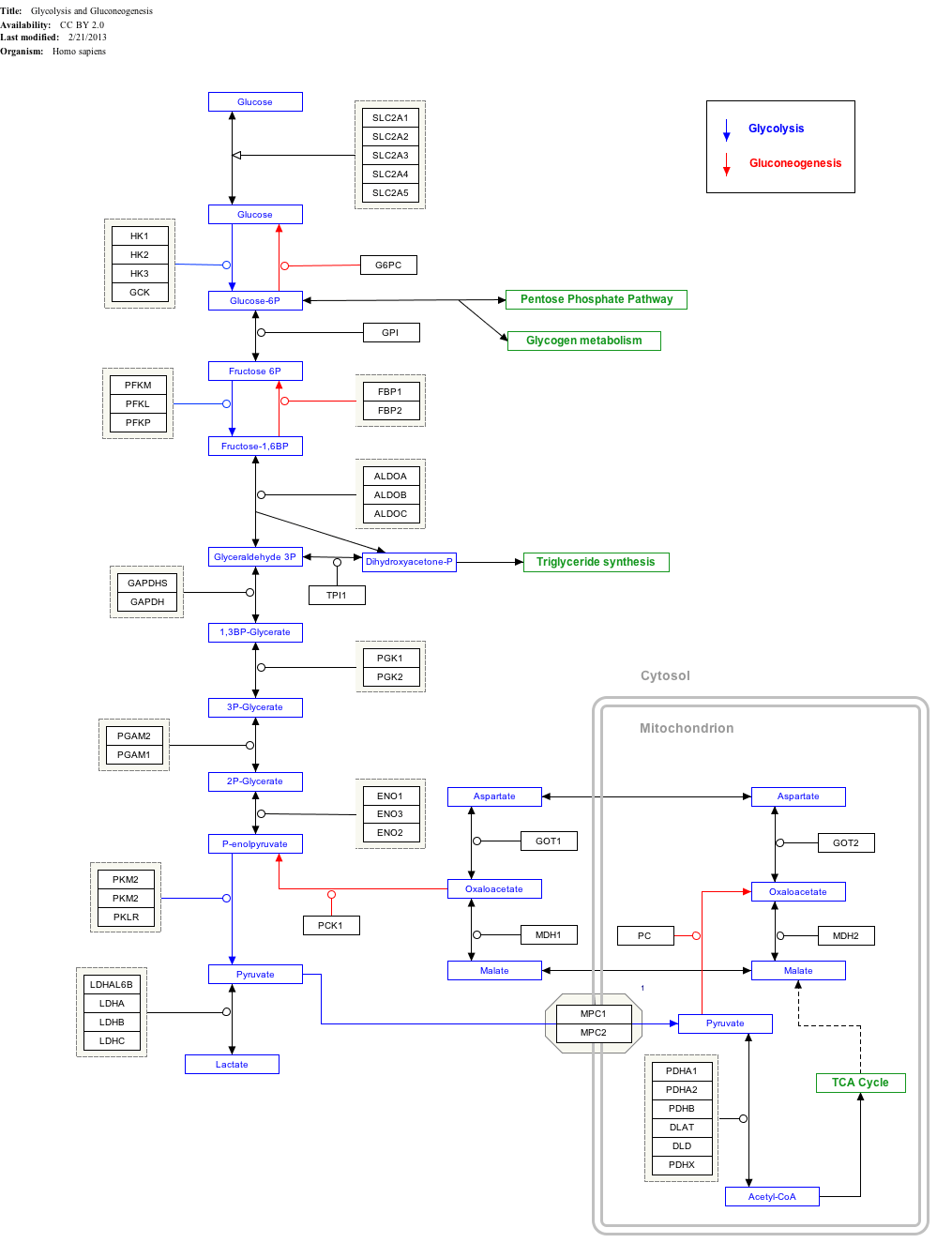
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
|{{{bSize}}}px|alt=Glycolysis and Gluconeogenesis edit]]
Glycolysis and Gluconeogenesis edit
- ↑ The interactive pathway map can be edited at WikiPathways: "GlycolysisGluconeogenesis_WP534".
Interactions
Alpha-enolase has been shown to interact with TRAPPC2.[26]
See also
External links
- Alpha-Enolase Linked to Severe Asthma - medscape news report, 25 aug 2006.
- Human ENO1 genome location and ENO1 gene details page in the UCSC Genome Browser.
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ "ENO1 enolase 1 (alpha)". NCBI Entrez Gene database.
- 1 2 3 4 5 6 7 8 Zhu X, Miao X, Wu Y, Li C, Guo Y, Liu Y, Chen Y, Lu X, Wang Y, He S (July 2015). "ENO1 promotes tumor proliferation and cell adhesion mediated drug resistance (CAM-DR) in Non-Hodgkin's Lymphomas". Experimental Cell Research. 335 (2): 216–23. PMID 26024773. doi:10.1016/j.yexcr.2015.05.020.
- ↑ Kim AY, Lim B, Choi J, Kim J (October 2016). "The TFG-TEC oncoprotein induces transcriptional activation of the human β-enolase gene via chromatin modification of the promoter region". Molecular Carcinogenesis. 55 (10): 1411–23. PMID 26310886. doi:10.1002/mc.22384.
- ↑ Giallongo A, Venturella S, Oliva D, Barbieri G, Rubino P, Feo S (June 1993). "Structural features of the human gene for muscle-specific enolase. Differential splicing in the 5'-untranslated sequence generates two forms of mRNA". European Journal of Biochemistry. 214 (2): 367–74. PMID 8513787. doi:10.1111/j.1432-1033.1993.tb17932.x.
- 1 2 3 4 5 6 7 8 9 Song Y, Luo Q, Long H, Hu Z, Que T, Zhang X, Li Z, Wang G, Yi L, Liu Z, Fang W, Qi S (March 2014). "Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma". Molecular Cancer. 13: 65. PMC 3994408
 . PMID 24650096. doi:10.1186/1476-4598-13-65.
. PMID 24650096. doi:10.1186/1476-4598-13-65. - 1 2 3 4 5 Subramanian A, Miller DM (February 2000). "Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene". The Journal of Biological Chemistry. 275 (8): 5958–65. PMID 10681589. doi:10.1074/jbc.275.8.5958.
- ↑ Subramanian A, Miller DM (February 2000). "Structural analysis of alpha-enolase. Mapping the functional domains involved in down-regulation of the c-myc protooncogene". The Journal of Biological Chemistry. 275 (8): 5958–65. PMID 10681589. doi:10.1074/jbc.275.8.5958.
- 1 2 3 4 Gao J, Zhao R, Xue Y, Niu Z, Cui K, Yu F, Zhang B, Li S (April 2013). "Role of enolase-1 in response to hypoxia in breast cancer: exploring the mechanisms of action". Oncology Reports. 29 (4): 1322–32. PMID 23381546. doi:10.3892/or.2013.2269.
- ↑ Pancholi V (June 2001). "Multifunctional alpha-enolase: its role in diseases". Cellular and Molecular Life Sciences. 58 (7): 902–20. PMID 11497239. doi:10.1007/pl00000910.
- 1 2 3 4 Hsiao KC, Shih NY, Fang HL, Huang TS, Kuo CC, Chu PY, Hung YM, Chou SW, Yang YY, Chang GC, Liu KJ (2013). "Surface α-enolase promotes extracellular matrix degradation and tumor metastasis and represents a new therapeutic target". PloS One. 8 (7): e69354. PMC 3716638
 . PMID 23894455. doi:10.1371/journal.pone.0069354.
. PMID 23894455. doi:10.1371/journal.pone.0069354. - 1 2 3 Chen S, Duan G, Zhang R, Fan Q (August 2014). "Helicobacter pylori cytotoxin-associated gene A protein upregulates α-enolase expression via Src/MEK/ERK pathway: implication for progression of gastric cancer". International Journal of Oncology. 45 (2): 764–70. PMID 24841372. doi:10.3892/ijo.2014.2444.
- ↑ Muller FL, Aquilanti EA, DePinho RA (November 2015). "Collateral Lethality: A new therapeutic strategy in oncology". Trends in Cancer. 1 (3): 161–173. PMID 26870836. doi:10.1016/j.trecan.2015.10.002.
- ↑ Duncan CG, Killela PJ, Payne CA, Lampson B, Chen WC, Liu J, Solomon D, Waldman T, Towers AJ, Gregory SG, McDonald KL, McLendon RE, Bigner DD, Yan H (August 2010). "Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes". Oncotarget. 1 (4): 265–77. PMID 21113414. doi:10.18632/oncotarget.137.
- ↑ Rus HG, Niculescu F, Vlaicu R (August 1991). "Tumor necrosis factor-alpha in human arterial wall with atherosclerosis". Atherosclerosis. 89 (2-3): 247–54. PMID 1793452. doi:10.1016/0021-9150(91)90066-C.
- 1 2 Leonard PG, Satani N, Maxwell D, Lin YH, Hammoudi N, Peng Z, Pisaneschi F, Link TM, Lee GR, Sun D, Prasad BA, Di Francesco ME, Czako B, Asara JM, Wang YA, Bornmann W, DePinho RA, Muller FL (December 2016). "SF2312 is a natural phosphonate inhibitor of enolase". Nature Chemical Biology. 12 (12): 1053–1058. PMID 27723749. doi:10.1038/nchembio.2195.
- 1 2 Muller FL, Colla S, Aquilanti E, Manzo VE, Genovese G, Lee J, Eisenson D, Narurkar R, Deng P, Nezi L, Lee MA, Hu B, Hu J, Sahin E, Ong D, Fletcher-Sananikone E, Ho D, Kwong L, Brennan C, Wang YA, Chin L, DePinho RA (August 2012). "Passenger deletions generate therapeutic vulnerabilities in cancer". Nature. 488 (7411): 337–42. PMID 22895339. doi:10.1038/nature11331.
- ↑ Moore TL, Gillian BE, Crespo-Pagnussat S, Feller L, Chauhan AK (2014). "Measurement and evaluation of isotypes of anti-citrullinated fibrinogen and anti-citrullinated alpha-enolase antibodies in juvenile idiopathic arthritis". Clinical and Experimental Rheumatology. 32 (5): 740–6. PMID 25068682.
- ↑ Yoneda M, Fujii A, Ito A, Yokoyama H, Nakagawa H, Kuriyama M (April 2007). "High prevalence of serum autoantibodies against the amino terminal of alpha-enolase in Hashimoto's encephalopathy". Journal of Neuroimmunology. 185 (1-2): 195–200. PMID 17335908. doi:10.1016/j.jneuroim.2007.01.018.
- ↑ Nahm DH, Lee KH, Shin JY, Ye YM, Kang Y, Park HS (August 2006). "Identification of alpha-enolase as an autoantigen associated with severe asthma". The Journal of Allergy and Clinical Immunology. 118 (2): 376–81. PMID 16890761. doi:10.1016/j.jaci.2006.04.002.
- ↑ Lee KH, Chung HS, Kim HS, Oh SH, Ha MK, Baik JH, Lee S, Bang D (July 2003). "Human alpha-enolase from endothelial cells as a target antigen of anti-endothelial cell antibody in Behçet's disease". Arthritis and Rheumatism. 48 (7): 2025–35. PMID 12847697. doi:10.1002/art.11074.
- ↑ Srivastava OP, Chandrasekaran D, Pfister RR (December 2006). "Molecular changes in selected epithelial proteins in human keratoconus corneas compared to normal corneas". Molecular Vision. 12: 1615–25. PMID 17200661.
- ↑ Nielsen K, Vorum H, Fagerholm P, Birkenkamp-Demtröder K, Honoré B, Ehlers N, Orntoft TF (February 2006). "Proteome profiling of corneal epithelium and identification of marker proteins for keratoconus, a pilot study". Experimental Eye Research. 82 (2): 201–9. PMID 16083875. doi:10.1016/j.exer.2005.06.009.
- ↑ Stefanini M (1972). "Chronic hemolytic anemia associated with erythrocyte enolase deficiency exacerbated by ingestion of nitrofurantoin". American Journal of Clinical Pathology. 58 (4): 408–14. PMID 4640298. doi:10.1093/ajcp/58.5.408.
- ↑ Ghosh AK, Majumder M, Steele R, White RA, Ray RB (January 2001). "A novel 16-kilodalton cellular protein physically interacts with and antagonizes the functional activity of c-myc promoter-binding protein 1". Molecular and Cellular Biology. 21 (2): 655–62. PMC 86643
 . PMID 11134351. doi:10.1128/MCB.21.2.655-662.2001.
. PMID 11134351. doi:10.1128/MCB.21.2.655-662.2001.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.