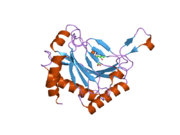EGLN1
Hypoxia-inducible factor prolyl hydroxylase 2 (HIF-PH2), or prolyl hydroxylase domain-containing protein 2 (PHD2), is an enzyme encoded by the EGLN1 gene. It is also known as Egl nine homolog 1.[3][4][5][6]
The hypoxia response
HIF-1α is a ubiquitous, constitutively synthesized transcription factor responsible for upregulating the expression of genes involved in the cellular response to hypoxia. These gene products may include proteins such as glycolytic enzymes and angiogenic growth factors.[7] In normoxia, HIF alpha subunits are marked for the ubiquitin-proteasome degradation pathway through hydroxylation of proline-564 and proline-402 by PHD2. Prolyl hydroxylation is critical for promoting pVHL binding to HIF, which targets HIF for polyubiquitylation.[6]
Structure
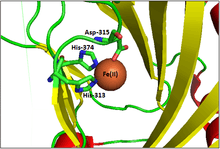
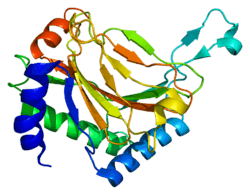
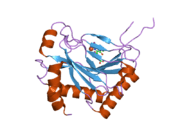
PHD2 is a 46-kDa enzyme that consists of an N-terminal domain homologous to MYND zinc finger domains, and a C-terminal domain homologous to the 2-oxoglutarate dioxygenases. The catalytic domain consists of a double-stranded β-helix core that is stabilized by three α-helices packed along the major β-sheet.[8] The active site, which is contained in the pocket between the β-sheets, chelates iron(II) through histidine and aspartate coordination. 2-oxoglutarate displaces a water molecule to bind iron as well.[9] The active site is lined by hydrophobic residues, possibly because such residues are less susceptible to potential oxidative damage by reactive species leaking from the iron center.[8]
The enzyme has a high affinity for iron(II) and 2-oxoglutarate, and forms a long-lived complex with these factors.[10] It has been proposed that cosubstrate and iron concentrations poise the HIF hydroxylases to respond to an appropriate "hypoxic window" for a particular cell type or tissue.[11] Studies have revealed that PHD2 has a KM for dioxygen slightly above its atmospheric concentration, and PHD2 is thought to be the most important sensor of the cell's oxygen status.[12]
Mechanism
The enzyme incorporates one oxygen atom from dioxygen into the hydroxylated product, and one oxygen atom into the succinate coproduct.[13] Its interactions with HIF-1α rely on a mobile loop region that helps to enclose the hydroxylation site and helps to stabilize binding of both iron and 2-oxyglutarate.[9]
_reaction_scheme.png)
Biological role and disease relevance
PHD2 is the primary regulator of HIF-1α steady state levels in the cell. A PHD2 knockdown showed increased levels of HIF-1α under normoxia, and an increase in HIF-1α nuclear accumulation and HIF-dependent transcription. HIF-1α steady state accumulation was dependent on the amount of PHD silencing effected by siRNA in HeLa cells and a variety of other human cell lines.[6]
However, although it would seem that PHD2 downregulates HIF-1α and thus also tumorigenesis, there have been suggestions of paradoxical roles of PHD2 in tumor proliferation. For example, one animal study showed tumor reduction in PHD2-deficient mice through activation of antiproliferative TGF-β signaling.[14] Other in vivo models showed tumor-suppressing activity for PHD2 in pancreatic cancer as well as liver cancer.[15][16] A study of 121 human patients revealed PHD2 as a strong prognostic marker in gastric cancer, with PHD2-negative patients having shortened survival compared to PHD2-positive patients.[17]
As an additional point of interest, recent genome-wide association studies have suggested that EGLN1 may be involved in the low hematocrit phenotype exhibited by the Tibetan population and hence that EGLN1 may play a role in the heritable adaptation of this population to live at high altitude.[18]
As a therapeutic target
HIF's important role as a homeostatic mediator implicates PHD2 as a therapeutic target for a range of disorders regarding angiogenesis, erythropoeisis, and cellular proliferation. There has been interest both in potentiating and inhibiting the activity of PHD2.[7] For example, methylselenocysteine (MSC) inhibition of HIF-1α led to tumor growth inhibition in renal cell carcinoma in a PHD-dependent manner. It is thought that this phenomenon relies on PHD-stabilization, but mechanistic details of this process have not yet been investigated.[19] On the other hand, screens of small-molecule chelators have revealed hydroxypyrones and hydroxypyridones as potential inhibitors for PHD2.[20] Substrate analog peptides have also been developed to exhibit inhibitory selectivity for PHD2 over factor inhibiting HIF (FIH), for which some other PHD-inhibitors show overlapping specificity.[21]
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- ↑ Dupuy D, Aubert I, Duperat VG, Petit J, Taine L, Stef M, Bloch B, Arveiler B (Nov 2000). "Mapping, characterization, and expression analysis of the SM-20 human homologue, c1orf12, and identification of a novel related gene, SCAND2". Genomics. 69 (3): 348–54. PMID 11056053. doi:10.1006/geno.2000.6343.
- ↑ Taylor MS (2001). "Characterization and comparative analysis of the EGLN gene family". Gene. 275 (1): 125–32. PMID 11574160. doi:10.1016/S0378-1119(01)00633-3.
- ↑ "Entrez Gene: EGLN1 egl nine homolog 1 (C. elegans)".
- 1 2 3 Berra E, Benizri E, Ginouvès A, Volmat V, Roux D, Pouysségur J (Aug 2003). "HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia". The EMBO Journal. 22 (16): 4082–4090. PMC 175782
 . PMID 12912907. doi:10.1093/emboj/cdg392.
. PMID 12912907. doi:10.1093/emboj/cdg392. - 1 2 William C, Nicholls L, Ratcliffe P, Pugh C, Maxwell P (2004). "The prolyl hydroxylase enzymes that act as oxygen sensors regulating destruction of hypoxia-inducible factor α". Advan. Enzyme Regul. 44: 75–92. PMID 15581484. doi:10.1016/j.advenzreg.2003.11.017.
- 1 2 McDonough M, Li V, Flashman E, Chowdhury R, Mohr C, Liénard ZJ, Oldham N, Clifton I, Lewis J, McNeill L, Kurzeja R, Hewitson K, Yang E, Jordan S, Syed R, Schofield C (Jun 2006). "Cellular oxygen sensing: Crystal structure of hypoxia-inducible factor prolyl hydroxylase (PHD2)". Proc Natl Acad Sci USA. 103 (26): 9814–9. PMC 1502536
 . PMID 16782814. doi:10.1073/pnas.0601283103.
. PMID 16782814. doi:10.1073/pnas.0601283103. - 1 2 Chowdhury R, McDonough M, Mecinović J, Loenarz C, Flashman E, Hewitson K, Domene C, Schofield C (Jul 2009). "Structural basis for binding of hypoxia-inducible factor to the oxygen-sensing prolyl hydroxylases". Structure. 17 (7): 981–9. PMID 19604478. doi:10.1016/j.str.2009.06.002.
- ↑ McNeill L, Flashman E, Buck M, Hewitson K, Clifton I, Jeschke G, Claridge T, Ehrismann D, Oldham N, Schofield C (Oct 2005). "Hypoxia-inducible factor prolyl hydroxylase 2 has a high affinity for ferrous iron and 2-oxoglutarate". Mol. Biosys. 1 (4): 321–4. PMID 16880998. doi:10.1039/b511249b.
- ↑ Ehrismann D, Flashman E, Genn DN, Mathioudakis N, Hewitson KS, Ratcliffe PJ, Schofield CJ (Jan 2007). "Studies on the activity of the hypoxia-inducible-factor hydroxylases using an oxygen consumption assay". Biochem. J. 401 (1): 227–34. PMC 1698668
 . PMID 16952279. doi:10.1042/BJ20061151.
. PMID 16952279. doi:10.1042/BJ20061151. - ↑ Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J (Aug 2003). "Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor". J. Biol. Chem. 278 (33): 30772–80. PMID 12788921. doi:10.1074/jbc.M304982200.
- ↑ McNeill LA, Hewitson KS, Gleadle JM, Horsfall LE, Oldham NJ, Maxwell PH, Pugh CW, Ratcliffe PJ, Schofield CJ (Jun 2002). "The use of dioxygen by HIF prolyl hydroxylase (PHD1)". Bioorg. Med. Chem. 12 (12): 1547–50. PMID 12039559. doi:10.1016/S0960-894X(02)00219-6.
- ↑ Ameln AK, Muschter A, Mamlouk S, Kalucka J, Prade I, Franke K, Rezaei M, Poitz DM, Breier G, Wielockx B (May 2011). "Inhibition of HIF prolyl hydroxylase-2 blocks tumor growth in mice through the antiproliferative activity of TGFβ". Cancer Res. 71 (9): 3306–16. PMID 21436457. doi:10.1158/0008-5472.CAN-10-3838.
- ↑ Su Y, Loos M, Giese N, Metzen E, Büchler MW, Friess H, Kornberg A, Büchler P (Feb 2012). "Prolyl hydroxylase-2 (PHD2) exerts tumor-suppressive activity in pancreatic cancer". Cancer. 118 (4): 960–72. PMID 21792862. doi:10.1002/cncr.26344.
- ↑ Heindryckx F, Kuchnio A, Casteleyn C, Coulon S, Olievier K, Colle I, Geerts A, Libbrecht L, Carmeliet P, Van Vlierberghe H (Jul 2012). "Effect of prolyl hydroxylase domain-2 haplodeficiency on the hepatocarcinogenesis in mice". J. Hepatol. 57 (1): 61–8. PMID 22420978. doi:10.1016/j.jhep.2012.02.021.
- ↑ Kamphues C, Wittschieber D, Klauschen F, Kasajima A, Dietel M, Schmidt SC, Glanemann M, Bahra M, Neuhaus P, Weichert W, Stenzinger A (Jan 2012). "Prolyl hydroxylase domain 2 protein is a strong prognostic marker in human gastric cancer". Pathobiology. 79 (1): 11–17. PMID 22236543. doi:10.1159/000330170.
- ↑ Simonson TS, Yang Y, Huff CD, Yun H, Qin G, Witherspoon DJ, Bai Z, Lorenzo FR, Xing J, Jorde LB, Prchal JT, Ge R (Jul 2010). "Genetic evidence for high-altitude adaptation in Tibet.". Science. 329 (5987): 72–5. PMID 20466884. doi:10.1126/science.1189406.
- ↑ Chintala S, Najrana T, Toth K, Cao S, Durrani F, Pili R, Rustum Y (2012). "Prolyl hydroxylase 2 dependent and Von-Hippel-Lindau independent degradation of hypoxia-inducible factor 1 and 2 alpha by selenium in clear cell renal cell carcinoma leads to tumor growth inhibition". BMC Cancer. 12: 293. PMC 3466155
 . PMID 22804960. doi:10.1186/1471-2407-12-293.
. PMID 22804960. doi:10.1186/1471-2407-12-293. - ↑ Flagg SC, Martin CB, Taabazuing CY, Holmes BE, Knapp MJ (Aug 2012). "Screening chelating inhibitors of HIF-prolyl hydroxylase domain 2 (PHD2) and factor inhibiting HIF (FIH)". J. Inorg. Biochem. 113: 25–30. PMC 3525482
 . PMID 22687491. doi:10.1016/j.jinorgbio.2012.03.002.
. PMID 22687491. doi:10.1016/j.jinorgbio.2012.03.002. - ↑ Kwon HS, Choi YK, Kim JW, Park YK, Yang EG, Ahn DR (Jul 2011). "Inhibition of a prolyl hydroxylase domain (PHD) by substrate analog peptides". Bioorg. Med. Chem. Lett. 21 (14): 4325–8. PMID 21665470. doi:10.1016/j.bmcl.2011.05.050.
Further reading
- Semenza GL (2001). "HIF-1, O(2), and the 3 PHDs: how animal cells signal hypoxia to the nucleus". Cell. 107 (1): 1–3. PMID 11595178. doi:10.1016/S0092-8674(01)00518-9.
- Wax SD, Tsao L, Lieb ME, et al. (1996). "SM-20 is a novel 40-kd protein whose expression in the arterial wall is restricted to smooth muscle". Lab. Invest. 74 (4): 797–808. PMID 8606489.
- Taylor MS (2001). "Characterization and comparative analysis of the EGLN gene family". Gene. 275 (1): 125–32. PMID 11574160. doi:10.1016/S0378-1119(01)00633-3.
- Epstein AC, Gleadle JM, McNeill LA, et al. (2001). "C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation". Cell. 107 (1): 43–54. PMID 11595184. doi:10.1016/S0092-8674(01)00507-4.
- Oehme F, Ellinghaus P, Kolkhof P, et al. (2002). "Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors". Biochem. Biophys. Res. Commun. 296 (2): 343–9. PMID 12163023. doi:10.1016/S0006-291X(02)00862-8.
- Ivan M, Haberberger T, Gervasi DC, et al. (2002). "Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor". Proc. Natl. Acad. Sci. U.S.A. 99 (21): 13459–64. PMC 129695
 . PMID 12351678. doi:10.1073/pnas.192342099.
. PMID 12351678. doi:10.1073/pnas.192342099. - Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. PMC 139241
 . PMID 12477932. doi:10.1073/pnas.242603899.
. PMID 12477932. doi:10.1073/pnas.242603899. - Metzen E, Berchner-Pfannschmidt U, Stengel P, et al. (2003). "Intracellular localisation of human HIF-1 alpha hydroxylases: implications for oxygen sensing". J. Cell. Sci. 116 (Pt 7): 1319–26. PMID 12615973. doi:10.1242/jcs.00318.
- Cioffi CL, Liu XQ, Kosinski PA, et al. (2003). "Differential regulation of HIF-1 alpha prolyl-4-hydroxylase genes by hypoxia in human cardiovascular cells". Biochem. Biophys. Res. Commun. 303 (3): 947–53. PMID 12670503. doi:10.1016/S0006-291X(03)00453-4.
- Aprelikova O, Chandramouli GV, Wood M, et al. (2004). "Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors". J. Cell. Biochem. 92 (3): 491–501. PMID 15156561. doi:10.1002/jcb.20067.
- Appelhoff RJ, Tian YM, Raval RR, et al. (2004). "Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor". J. Biol. Chem. 279 (37): 38458–65. PMID 15247232. doi:10.1074/jbc.M406026200.
- Metzen E, Stiehl DP, Doege K, et al. (2006). "Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element". Biochem. J. 387 (Pt 3): 711–7. PMC 1135001
 . PMID 15563275. doi:10.1042/BJ20041736.
. PMID 15563275. doi:10.1042/BJ20041736. - Baek JH, Mahon PC, Oh J, et al. (2005). "OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1alpha". Mol. Cell. 17 (4): 503–12. PMID 15721254. doi:10.1016/j.molcel.2005.01.011.
- Ozer A, Wu LC, Bruick RK (2005). "The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF)". Proc. Natl. Acad. Sci. U.S.A. 102 (21): 7481–6. PMC 1140452
 . PMID 15897452. doi:10.1073/pnas.0502716102.
. PMID 15897452. doi:10.1073/pnas.0502716102. - Choi KO, Lee T, Lee N, et al. (2006). "Inhibition of the catalytic activity of hypoxia-inducible factor-1alpha-prolyl-hydroxylase 2 by a MYND-type zinc finger". Mol. Pharmacol. 68 (6): 1803–9. PMID 16155211. doi:10.1124/mol.105.015271.
- To KK, Huang LE (2006). "SUPPRESSION OF HIF-1α TRANSCRIPTIONAL ACTIVITY BY THE HIF PROLYL HYDROXYLASE EGLN1". J. Biol. Chem. 280 (45): 38102–7. PMC 1307502
 . PMID 16157596. doi:10.1074/jbc.M504342200.
. PMID 16157596. doi:10.1074/jbc.M504342200. - Kato H, Inoue T, Asanoma K, et al. (2006). "Induction of human endometrial cancer cell senescence through modulation of HIF-1alpha activity by EGLN1". Int. J. Cancer. 118 (5): 1144–53. PMID 16161047. doi:10.1002/ijc.21488.