CrcB RNA motif
| crcB RNA motif | |
|---|---|
 Consensus secondary structure of crcB RNAs | |
| Identifiers | |
| Symbol | crcB RNA |
| Rfam | RF01734 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Prokaryota |
The crcB RNA motif (now called the fluoride riboswitch) is a conserved RNA structure identified by bioinformatics in a wide variety of bacteria and archaea.[1] These RNAs were later shown to function as riboswitches that sense fluoride ions.[2] These "fluoride riboswitches" increase expression of downstream genes when fluoride levels are elevated, and the genes are proposed to help mitigate the toxic effects of very high levels of fluoride.
Many genes are presumed to be regulated by these fluoride riboswitches. Two of the most common encode proteins that are proposed to function by removing fluoride from the cell. These proteins are CrcB proteins and a fluoride-specific subtype of chloride channels referred to as EriC^F or ClC^F. ClC^F proteins have been shown to function as fluoride-specific fluoride/proton antiporters.[3]
The three-dimensional structure of a fluoride riboswitch has been solved at atomic resolution by X-ray crystallography.[4]
Fluoride riboswitches are found in many organisms within the domains bacteria and archaea, indicating that many of these organisms sometimes encounter elevated levels of fluoride. Of particular interest is Streptococcus mutans, a major cause of dental caries. It has been shown that sodium fluoride has inhibited the growth rate of S. mutans using glucose as an energy and carbon source.[5] However, it is also noteworthy that many organisms that do not encounter fluoride in the human mouth carry fluoride riboswitches or resistance genes.
Discovery of the Fluoride Riboswitch
The identity of fluoride as the riboswitch ligand was accidentally discovered when a compound contaminated with fluoride caused significant conformational changes to the non-coding crcB RNA motif during an in-line probing experiment.[2] In-line probing was used to illuminate the secondary structure of the crcB RNA motif and structural changes associated with possible binding to specific metabolites or ions.[6] The results of the probing showed the addition of increasing fluoride ion concentrations suppressed certain regions of spontaneous RNA cleavage and heightening other regions. These nucleotide regions in the crcB RNA motif play important roles in the aptamer binding region for fluoride.[2]
Upon binding fluoride ions, the fluoride riboswitch showed regulation of downstream gene transcription.[2] These downstream genes transcribe fluoride sensitive enzymes [2] such as enolase, pyrophosphatase, the presumed fluoride exporter CrcB and a superfamily of CLC membrane proteins called Eric^F proteins.[3] The CLC^F proteins have been shown to function as fluoride transporters against fluoride toxicity.[3] The eric^F gene is a mutant version of the chloride channel gene that is less common in bacteria than chloride-specific homologs, but is nonetheless found in the genome of Streptococcus mutans.[7] The Eric^F protein in particular carries specific amino acids in their channels that targets fluoride anions whereas the regular Eric protein favored chloride over fluoride ions.[2]
Fluoride Riboswitch Structure
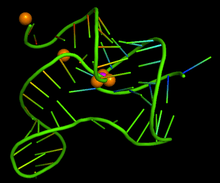
The discovery of the fluoride riboswitch was surprising as both fluoride ions and the crcB RNA phosphate groups are negatively charged and should not be able to bind to one another.[2] Previous research came across this question in elucidating the cofactor thiamine pyrophosphate (TPP) riboswitch. The TPP riboswitch structure showed the assistance of two hydrated Mg^2+ ions that help stabilize the connection between the phosphates of TPP and guanine bases of the RNA.[8][9] This guiding research help characterize the fluoride riboswitch’s own interactions with fluoride and its structure. Through in-line probing and mutational studies the fluoride riboswitch of the organism Thermotoga petrophila is recognized to have two helical stems adjoined by a helical loop with the capacity to become a pseudoknot.[10] The bound fluoride ligand is found to be located within the center of the riboswitch fold, enclosed by three Mg^2+ ions. The Mg^2+ ions are octahedrally coordinated with five outer backbone phosphates and water molecules making a metabolite specific pocket for coordinating the fluoride ligand to bind. The placement of the Mg^2+ ions positions the fluoride ion into the negatively charged crcB RNA scaffold.[10]
Biological Significance

In the earth’s crust, fluoride is the 13th most abundant element.[2] It is commonly used in oral healthcare products and water.[2] The fluoride acts as a hardening agent with the enamel base on teeth, remineralizing and protecting them from harsh acids and bacteria in the oral cavity.[11] Additionally, its significance lies in the effect of the toxicity of fluoride at high concentrations to bacteria, especially those that cause dental caries. It has long been known that many species encapsulate a sensor system for toxic metals such as cadmium and silver.[2] However, a sensor system against fluoride remained unknown. The fluoride riboswitch elucidates the bacterial defense mechanism in counteracting against the toxicity of high concentrations of fluoride by regulating downstream genes of the riboswitch upon binding the fluoride ligand.[2] Further elucidating the mechanism of how bacteria protect itself from fluoride toxicity can help modify the mechanism to make smaller concentrations of fluoride even more lethal to bacteria. Additionally, the fluoride riboswitch and the downstream regulated genes can be potential targets for drug development in the future. Overall, these advancements will help towards making fluoride and future drugs strong protectors against oral health disease.
References
- ↑ Weinberg Z, Wang JX, Bogue J, et al. (March 2010). "Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea and their metagenomes". Genome Biol. 11 (3): R31. PMC 2864571
 . PMID 20230605. doi:10.1186/gb-2010-11-3-r31.
. PMID 20230605. doi:10.1186/gb-2010-11-3-r31. - 1 2 3 4 5 6 7 8 9 10 11 Baker JL, Sudarsan N, Weinberg Z, et al. (January 2012). "Widespread genetic switches and toxicity resistance proteins for fluoride". Science. 335 (6065): 233–5. PMID 22194412. doi:10.1126/science.1215063.
- 1 2 3 Stockbridge, RB; Lim HH; Otten R; Williams C; Shane T; Weinberg Z; Miller C (18 September 2012). "Fluoride resistance and transport by riboswitch-controlled CLC antiporters.". Proc Natl Acad Sci U S A. 109 (38): 15289–94. PMC 3458365
 . PMID 22949689. doi:10.1073/pnas.1210896109.
. PMID 22949689. doi:10.1073/pnas.1210896109. - ↑ Ren A, Rajashankar KR, Patel DJ (June 2012). "Fluoride ion encapsulation by Mg2+ ions and phosphates in a fluoride riboswitch". Nature. 486 (7401): 85–9. PMC 3744881
 . PMID 22678284. doi:10.1038/nature11152.
. PMID 22678284. doi:10.1038/nature11152. - ↑ Yost, K G; VanDemark, P J (May 1978). "Growth inhibition of Streptococcus mutans and Leuconostoc mesenteroides by sodium fluoride and ionic tin". Applied and Environmental Microbiology. 35: 920–924. PMC 242953
 . PMID 655708.
. PMID 655708. - ↑ Regulski, EE; Breaker RR (2008). "In-line probing analysis of riboswitches". Methods molecular biology. 419: 53–67. PMID 18369975. doi:10.1007/978-1-59745-033-1_4.
- ↑ Breaker, R.R. (10 February 2012). "New Insight on the Response of Bacteria to Fluoride". Caries Research. 46: 78–81. doi:10.1159/000336397. Retrieved 25 February 2013.
- ↑ Serganov, A; Polonskaia A; Phan AT; Breaker RR; Patel DJ (29 June 2006). "Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch.". Nature. 441 (7097): 1167–71. PMID 16728979. doi:10.1038/nature04740.
- ↑ Thore, S; Leibundgut M; Ban N (26 May 2006). "Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand.". Science. 312 (5777): 1208–11. PMID 16675665. doi:10.1126/science.1128451.
- 1 2 Ren, A; Rajashankar KR; Patel DJ (13 May 2012). "Fluoride ion encapsulation by Mg^2+ ions and phosphates in a fluoride riboswitch.". Nature. 486 (7401): 85–9. PMC 3744881
 . PMID 22678284. doi:10.1038/nature11152.
. PMID 22678284. doi:10.1038/nature11152. - ↑ Wolfgang, Arnold; Andreas Dorow; Stephanie Langenhorst; Zeno Gintner; Jolan Banoczy; Peter Gaengler (15 June 2006). "Effect of fluoride toothpastes on enamel demineralization". BMC Oral Health. 6 (8). PMC 1543617
 . PMID 16776820. doi:10.1186/1472-6831-6-8.
. PMID 16776820. doi:10.1186/1472-6831-6-8.