Central dogma of molecular biology
The central dogma of molecular biology is an explanation of the flow of genetic information within a biological system. It was first stated by Francis Crick in 1958[1]
| “ | The Central Dogma. This states that once ‘information’ has passed into protein it cannot get out again. In more detail, the transfer of information from nucleic acid to nucleic acid, or from nucleic acid to protein may be possible, but transfer from protein to protein, or from protein to nucleic acid is impossible. Information means here the precise determination of sequence, either of bases in the nucleic acid or of amino acid residues in the protein. | ” |
| — Francis Crick, 1958 | ||
and re-stated in a Nature paper published in 1970:[2]
| “ | The central dogma of molecular biology deals with the detailed residue-by-residue transfer of sequential information. It states that such information cannot be transferred back from protein to either protein or nucleic acid. | ” |
| — Francis Crick | ||
The central dogma has also been described as "DNA makes RNA and RNA makes protein,"[3] originally termed the sequence hypothesis and made as a positive statement by Crick. However, this simplification does not make it clear that the central dogma as stated by Crick does not preclude the reverse flow of information from RNA to DNA, only ruling out the flow from protein to RNA or DNA. Crick's use of the word dogma was unconventional, and has been controversial.
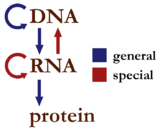
The dogma is a framework for understanding the transfer of sequence information between information-carrying biopolymers, in the most common or general case, in living organisms. There are 3 major classes of such biopolymers: DNA and RNA (both nucleic acids), and protein. There are 3×3 = 9 conceivable direct transfers of information that can occur between these. The dogma classes these into 3 groups of 3: three general transfers (believed to occur normally in most cells), three special transfers (known to occur, but only under specific conditions in case of some viruses or in a laboratory), and three unknown transfers (believed never to occur). The general transfers describe the normal flow of biological information: DNA can be copied to DNA (DNA replication), DNA information can be copied into mRNA (transcription), and proteins can be synthesized using the information in mRNA as a template (translation). The special transfers describe: RNA being copied from RNA (RNA replication), DNA being synthesised using an RNA template (reverse transcription), and proteins being synthesised directly from a DNA template without the use of mRNA. The unknown transfers describe: a protein being copied from a protein, synthesis of RNA using the primary structure of a protein as a template, and DNA synthesis using the primary structure of a protein as a template - these are not thought to naturally occur.[2]
Biological sequence information
The biopolymers that comprise DNA, RNA and (poly)peptides are linear polymers (i.e.: each monomer is connected to at most two other monomers). The sequence of their monomers effectively encodes information. The transfers of information described by the central dogma ideally are faithful, deterministic transfers, wherein one biopolymer's sequence is used as a template for the construction of another biopolymer with a sequence that is entirely dependent on the original biopolymer's sequence.
General transfers of biological sequential information

Table of the three classes of information transfer suggested by the dogma General Special Unknown DNA → DNA RNA → DNA protein → DNA DNA → RNA RNA → RNA protein → RNA RNA → protein DNA → protein protein → protein
DNA replications
In the sense that DNA replication must occur if genetic material is to be provided for the progeny of any cell, whether somatic or reproductive, the copying from DNA to DNA arguably is the fundamental step in the central dogma. A complex group of proteins called the replisome performs the replication of the information from the parent strand to the complementary daughter strand.
The replisome comprises:
- a helicase that unwinds the superhelix as well as the double-stranded DNA helix to create a replication fork
- SSB protein that binds open the double-stranded DNA to prevent it from reassociating
- RNA primase that adds a complementary RNA primer to each template strand as a starting point for replication
- DNA polymerase III that reads the existing template chain from its 3' end to its 5' end and adds new complementary nucleotides from the 5' end to the 3' end of the daughter chain
- DNA polymerase I that removes the RNA primers and replaces them with DNA.
- DNA ligase that joins the two Okazaki fragments with phosphodiester bonds to produce a continuous chain.
This process typically takes place during S phase of the cell cycle.
Transcription

Transcription is the process by which the information contained in a section of DNA is replicated in the form of a newly assembled piece of messenger RNA (mRNA). Enzymes facilitating the process include RNA polymerase and transcription factors. In eukaryotic cells the primary transcript is pre-mRNA. Pre-mRNA must be processed for translation to proceed. Processing includes the addition of a 5' cap and a poly-A tail to the pre-mRNA chain, followed by splicing. Alternative splicing occurs when appropriate, increasing the diversity of the proteins that any single mRNA can produce. The product of the entire transcription process (that began with the production of the pre-mRNA chain) is a mature mRNA chain.
Translation
The mature mRNA finds its way to a ribosome, where it gets translated. In prokaryotic cells, which have no nuclear compartment, the processes of transcription and translation may be linked together without clear separation. In eukaryotic cells, the site of transcription (the cell nucleus) is usually separated from the site of translation (the cytoplasm), so the mRNA must be transported out of the nucleus into the cytoplasm, where it can be bound by ribosomes. The ribosome reads the mRNA triplet codons, usually beginning with an AUG (adenine−uracil−guanine), or initiator methionine codon downstream of the ribosome binding site. Complexes of initiation factors and elongation factors bring aminoacylated transfer RNAs (tRNAs) into the ribosome-mRNA complex, matching the codon in the mRNA to the anti-codon on the tRNA. Each tRNA bears the appropriate amino acid residue to add to the polypeptide chain being synthesised. As the amino acids get linked into the growing peptide chain, the chain begins folding into the correct conformation. Translation ends with a stop codon which may be a UAA, UGA, or UAG triplet.
The mRNA does not contain all the information for specifying the nature of the mature protein. The nascent polypeptide chain released from the ribosome commonly requires additional processing before the final product emerges. For one thing, the correct folding process is complex and vitally important. For most proteins it requires other chaperone proteins to control the form of the product. Some proteins then excise internal segments from their own peptide chains, splicing the free ends that border the gap; in such processes the inside "discarded" sections are called inteins. Other proteins must be split into multiple sections without splicing. Some polypeptide chains need to be cross-linked, and others must be attached to cofactors such as haem (heme) before they become functional.
Special transfers of biological sequential information
Reverse transcription
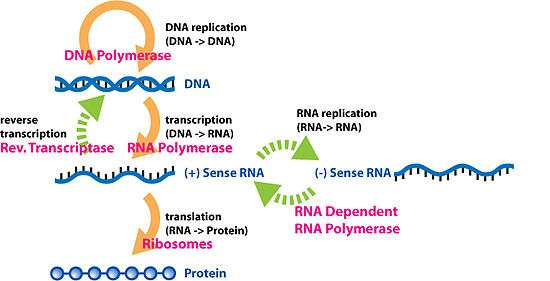
Reverse transcription is the transfer of information from RNA to DNA (the reverse of normal transcription). This is known to occur in the case of retroviruses, such as HIV, as well as in eukaryotes, in the case of retrotransposons and telomere synthesis. It is the process by which genetic information from RNA gets transcribed into new DNA.
RNA replication
RNA replication is the copying of one RNA to another. Many viruses replicate this way. The enzymes that copy RNA to new RNA, called RNA-dependent RNA polymerases, are also found in many eukaryotes where they are involved in RNA silencing.[4]
RNA editing, in which an RNA sequence is altered by a complex of proteins and a "guide RNA", could also be seen as an RNA-to-RNA transfer.
Direct translation from DNA to protein
Direct translation from DNA to protein has been demonstrated in a cell-free system (i.e. in a test tube), using extracts from E. coli that contained ribosomes, but not intact cells. These cell fragments could synthesize proteins from single-stranded DNA templates isolated from other organisms (e,g., mouse or toad), and neomycin was found to enhance this effect. However, it was unclear whether this mechanism of translation corresponded specifically to the genetic code.[5][6]
Transfers of information not explicitly covered in the theory
Posttranslational modification
After protein amino acid sequences have been translated from nucleic acid chains, they can be edited by appropriate enzymes. Although this is a form of protein affecting protein sequence, not explicitly covered by the central dogma, there are not many clear examples where the associated concepts of the two fields have much to do with each other.
Inteins
An intein is a "parasitic" segment of a protein that is able to excise itself from the chain of amino acids as they emerge from the ribosome and rejoin the remaining portions with a peptide bond in such a manner that the main protein "backbone" does not fall apart. This is a case of a protein changing its own primary sequence from the sequence originally encoded by the DNA of a gene. Additionally, most inteins contain a homing endonuclease or HEG domain which is capable of finding a copy of the parent gene that does not include the intein nucleotide sequence. On contact with the intein-free copy, the HEG domain initiates the DNA double-stranded break repair mechanism. This process causes the intein sequence to be copied from the original source gene to the intein-free gene. This is an example of protein directly editing DNA sequence, as well as increasing the sequence's heritable propagation.
Methylation
Variation in methylation states of DNA can alter gene expression levels significantly. Methylation variation usually occurs through the action of DNA methylases. When the change is heritable, it is considered epigenetic. When the change in information status is not heritable, it would be a somatic epitype. The effective information content has been changed by means of the actions of a protein or proteins on DNA, but the primary DNA sequence is not altered.
Prions
Prions are proteins of particular amino acid sequences in particular conformations. They propagate themselves in host cells by making conformational changes in other molecules of protein with the same amino acid sequence, but with a different conformation that is functionally important or detrimental to the organism. Once the protein has been transconformed to the prion folding it changes function. In turn it can convey information into new cells and reconfigure more functional molecules of that sequence into the alternate prion form. In some types of prion in fungi this change is continuous and direct; the information flow is Protein → Protein.
Some scientists such as Alain E. Bussard and Eugene Koonin have argued that prion-mediated inheritance violates the central dogma of molecular biology.[7][8] However, Rosalind Ridley in Molecular Pathology of the Prions (2001) has written that "The prion hypothesis is not heretical to the central dogma of molecular biology—that the information necessary to manufacture proteins is encoded in the nucleotide sequence of nucleic acid—because it does not claim that proteins replicate. Rather, it claims that there is a source of information within protein molecules that contributes to their biological function, and that this information can be passed on to other molecules."[9]
Natural genetic engineering
James A. Shapiro argues that a superset of these examples should be classified as natural genetic engineering and are sufficient to falsify the central dogma. While Shapiro has received a respectful hearing for his view, his critics have not been convinced that his reading of the central dogma is in line with what Crick intended.[10] [11]
Use of the term "dogma"
In his autobiography, What Mad Pursuit, Crick wrote about his choice of the word dogma and some of the problems it caused him:
"I called this idea the central dogma, for two reasons, I suspect. I had already used the obvious word hypothesis in the sequence hypothesis, and in addition I wanted to suggest that this new assumption was more central and more powerful. ... As it turned out, the use of the word dogma caused almost more trouble than it was worth. Many years later Jacques Monod pointed out to me that I did not appear to understand the correct use of the word dogma, which is a belief that cannot be doubted. I did apprehend this in a vague sort of way but since I thought that all religious beliefs were without foundation, I used the word the way I myself thought about it, not as most of the world does, and simply applied it to a grand hypothesis that, however plausible, had little direct experimental support."
Similarly, Horace Freeland Judson records in The Eighth Day of Creation:[12]
"My mind was, that a dogma was an idea for which there was no reasonable evidence. You see?!" And Crick gave a roar of delight. "I just didn't know what dogma meant. And I could just as well have called it the 'Central Hypothesis,' or — you know. Which is what I meant to say. Dogma was just a catch phrase."
See also
References
- ↑ Crick, F.H.C. (1958). "On Protein Synthesis". In F.K. Sanders. Symposia of the Society for Experimental Biology, Number XII: The Biological Replication of Macromolecules. Cambridge University Press. pp. 138–163.
- 1 2 Crick, F (August 1970). "Central dogma of molecular biology." (PDF). Nature. 227 (5258): 561–3. Bibcode:1970Natur.227..561C. PMID 4913914. doi:10.1038/227561a0.
- ↑ Leavitt, Sarah A. (June 2010). "Deciphering the Genetic Code: Marshall Nirenberg". Office of NIH History.
- ↑ Ahlquist P (May 2002). "RNA-dependent RNA polymerases, viruses, and RNA silencing". Science. 296 (5571): 1270–3. Bibcode:2002Sci...296.1270A. PMID 12016304. doi:10.1126/science.1069132.
- ↑ B. J. McCarthy; J. J. Holland (September 15, 1965). "Denatured DNA as a Direct Template for in vitro Protein Synthesis". Proceedings of the National Academy of Sciences of the United States of America. 54 (3): 880–886. Bibcode:1965PNAS...54..880M. PMC 219759
 . PMID 4955657. doi:10.1073/pnas.54.3.880.
. PMID 4955657. doi:10.1073/pnas.54.3.880. - ↑ .T. Uzawa; A. Yamagishi; T. Oshima (2002-04-09). "Polypeptide Synthesis Directed by DNA as a Messenger in Cell-Free Polypeptide Synthesis by Extreme Thermophiles, Thermus thermophilus HB27 and Sulfolobus tokodaii Strain 7". The Journal of Biochemistry. 131 (6): 849–853. PMID 12038981. doi:10.1093/oxfordjournals.jbchem.a003174.
- ↑ Bussard Alain E (2005). "A scientific revolution? The prion anomaly may challenge the central dogma of molecular biology". EMBO Rep. 6 (8): 691–694. PMC 1369155
 . PMID 16065057. doi:10.1038/sj.embor.7400497.
. PMID 16065057. doi:10.1038/sj.embor.7400497. - ↑ Koonin, Eugene. (2012). Does the Central Dogma Still Stand? Biology Direct 7: 27.
- ↑ Ridley, Rosalind. (2001). What Would Thomas Henry Huxley Have Made of Prion Diseases?. In Harry F. Baker. Molecular Pathology of the Prions (Methods in Molecular Medicine). Humana Press. pp. 1-16. ISBN 0-89603-924-2
- ↑ Wilkins, Adam S. (January 2012). "(Review) Evolution: A View from the 21st Century". Genome Biology and Evolution. 4: 423–426. doi:10.1093/gbe/evs008.
- ↑ Moran, Laurence A (May–June 2011). "(Review) Evolution: A View from the 21st Century". Reports of the National Center for Science Education. 32.3 (9): 1–4.
- ↑ Horace Freeland Judson (1996). "Chapter 6: My mind was, that a dogma was an idea for which there was no reasonable evidence. You see?!". The Eighth Day of Creation: Makers of the Revolution in Biology (25th anniversary edition). Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. ISBN 0-87969-477-7.
Further reading
- Bussard Alain E (2005). "A scientific revolution? The prion anomaly may challenge the central dogma of molecular biology". EMBO Rep. 6 (8): 691–694. PMC 1369155
 . PMID 16065057. doi:10.1038/sj.embor.7400497.
. PMID 16065057. doi:10.1038/sj.embor.7400497. - Baker, Harry F. (2001). Molecular Pathology of the Prions (Methods in Molecular Medicine). Humana Press. ISBN 0-89603-924-2
- Koonin Eugene (2012). "Does the Central Dogma Still Stand?". Biology Direct. 7: 27. doi:10.1186/1745-6150-7-27.
- Li J. J; Biggin M. D. (2015). "Gene expression. Statistics requantitates the central dogma". Science. 347 (6226): 1066–1067. PMID 25745146. doi:10.1126/science.aaa8332.
- Piras V, Tomita M, Selvarajoo K (2012). "Is central dogma a global property of cellular information flow?". Frontiers in Physiology. 3: 439. doi:10.3389/fphys.2012.00439.
- Robinson Victoria L (2009). "Rethinking the Central Dogma: noncoding RNAs are biologically relevant". Urologic Oncology. 27 (3): 304–306. doi:10.1016/j.urolonc.2008.11.004.
External links
| Wikimedia Commons has media related to Central dogma of molecular biology. |
- The Elaboration of the Central Dogma – Scitable: By Nature education
- Animation of Central Dogma from RIKEN - NatureDocumentaries.org
- Discussion on challenges to the "Central Dogma of Molecular Biology"
- Explanation of the central dogma using a musical analogy
- "Francis Harry Compton Crick (1916–2004)" by A. Andrei at the Embryo Project Encyclopedia