Pyrosequencing

Pyrosequencing is a method of DNA sequencing (determining the order of nucleotides in DNA) based on the "sequencing by synthesis" principle. It differs from Sanger sequencing, in that it relies on the detection of pyrophosphate release on nucleotide incorporation, rather than chain termination with dideoxynucleotides.[1] The technique was developed by Mostafa Ronaghi and Pål Nyrén at the Royal Institute of Technology in Stockholm in 1996.[2][3][4] The desired DNA sequence is able to be determined by light emitted upon incorporation of the next complementary nucleotide by the fact that only one out of four of the possible A/T/C/G nucleotides are added and available at a time so that only one letter can be incorporated on the single stranded template (which is the sequence to be determined). The intensity of the light determines if there are more than one of these "letters" in a row. The previous nucleotide letter (one out of four possible dNTP) is degraded before the next nucleotide letter is added for synthesis: allowing for the possible revealing of the next nucleotide(s) via the resulting intensity of light (if the nucleotide added was the next complementary letter in the sequence). This process is repeated with each of the four letters until the DNA sequence of the single stranded template is determined.
==Procedure== [5]
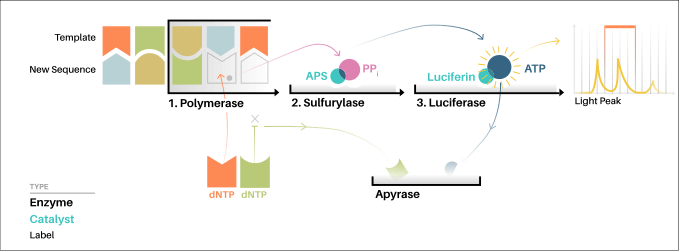
"Sequencing by synthesis" involves taking a single strand of the DNA to be sequenced and then synthesizing its complementary strand enzymatically. The pyrosequencing method is based on detecting the activity of DNA polymerase (a DNA synthesizing enzyme) with another chemoluminescent enzyme. Essentially, the method allows sequencing of a single strand of DNA by synthesizing the complementary strand along it, one base pair at a time, and detecting which base was actually added at each step. The template DNA is immobile, and solutions of A, C, G, and T nucleotides are sequentially added and removed from the reaction. Light is produced only when the nucleotide solution complements the first unpaired base of the template. The sequence of solutions which produce chemiluminescent signals allows the determination of the sequence of the template.
The single-strand DNA (ssDNA) template is hybridized to a sequencing primer and incubated with the enzymes DNA polymerase, ATP sulfurylase, luciferase and apyrase, and with the substrates adenosine 5´ phosphosulfate (APS) and luciferin.
- The addition of one of the four deoxynucleotide triphosphates (dNTPs) (dATPαS, which is not a substrate for a luciferase, is added instead of dATP to avoid noise) initiates the second step. DNA polymerase incorporates the correct, complementary dNTPs onto the template. This incorporation releases pyrophosphate (PPi).
- ATP sulfurylase converts PPi to ATP in the presence of adenosine 5´ phosphosulfate. This ATP acts as a substrate for the luciferase-mediated conversion of luciferin to oxyluciferin that generates visible light in amounts that are proportional to the amount of ATP. The light produced in the luciferase-catalyzed reaction is detected by a camera and analyzed in a pyrogram.
- Unincorporated nucleotides and ATP are degraded by the apyrase, and the reaction can restart with another nucleotide.
Currently, a limitation of the method is that the lengths of individual reads of DNA sequence are in the neighborhood of 300-500 nucleotides, shorter than the 800-1000 obtainable with chain termination methods (e.g. Sanger sequencing). This can make the process of genome assembly more difficult, particularly for sequences containing a large amount of repetitive DNA. As of 2007, pyrosequencing is most commonly used for resequencing or sequencing of genomes for which the sequence of a close relative is already available.
The templates for pyrosequencing can be made both by solid phase template preparation (streptavidin-coated magnetic beads) and enzymatic template preparation (apyrase+exonuclease). So Pyrosequencing can be differentiated into two types, namely Solid Phase Pyrosequencing and Liquid Phase Pyrosequencing.
Commercialization
The company Pyrosequencing AB in Uppsala, Sweden was founded with venture capital provided by HealthCap in order to commercialize machinery and reagents for sequencing short stretches of DNA using the pyrosequencing technique. Pyrosequencing AB was listed on the Stockholm Stock Exchange in 1999. It was renamed to Biotage in 2003. The pyrosequencing business line was acquired by Qiagen in 2008.[6] Pyrosequencing technology was further licensed to 454 Life Sciences. 454 developed an array-based pyrosequencing technology which has emerged as a platform for large-scale DNA sequencing. Most notable are the applications for genome sequencing and metagenomics. GS FLX, the latest pyrosequencing platform by 454 Life Sciences (now owned by Roche Diagnostics), can generate 400 Mb in a 10-hour run with a single machine. Each run would cost about 5,000-7,000 USD.
Hoffman LaRoche announced the discontinuation of the 454 sequencing platform in 2013.
References
- ↑ "Definition of pyrosequencing from the Nature Reviews Genetics Glossary". Retrieved 2008-10-28.
- ↑ Ronaghi; Uhlén, M; Nyrén, P (1998-07-17). "A sequencing method based on real-time pyrophosphate". Science. 281 (5375): 363. PMID 9705713. doi:10.1126/science.281.5375.363.
- ↑ Ronaghi; Karamohamed, S; Pettersson, B; Uhlén, M; Nyrén, P (1996). "Real-time DNA sequencing using detection of pyrophosphate release". Analytical Biochemistry. 242 (1): 84–9. PMID 8923969. doi:10.1006/abio.1996.0432.
- ↑ Nyrén, P. (2007). "The History of Pyrosequencing". Methods Mol Biology. 373: 1–14. PMID 17185753. doi:10.1385/1-59745-377-3:1.
- ↑ QIAGEN. "Pyrosequencing Technology and Platform Overview". Retrieved 4 August 2017.
- ↑ Qiagen acquires Biotage
Further reading
- Metzker M. (2005). "Emerging Technologies in DNA Sequencing". Genome Research. 15 (12): 1767–76. PMID 16339375. doi:10.1101/gr.3770505.