ATP synthase
| ATP Synthase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
|
Molecular model of ATP synthase determined by X-ray crystallography | |||||||||
| Identifiers | |||||||||
| EC number | 3.6.3.14 | ||||||||
| CAS number | 9000-83-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / EGO | ||||||||
| |||||||||
ATP synthase is an enzyme that creates the energy storage molecule adenosine triphosphate (ATP). ATP is the most commonly used "energy currency" of cells for most organisms. It is formed from adenosine diphosphate (ADP) and inorganic phosphate (Pi). The overall reaction catalyzed by ATP synthase is:
- ADP + Pi + H+out ⇌ ATP + H2O + H+in
The formation of ATP from ADP and Pi is energetically unfavorable and would normally proceed in the reverse direction. In order to drive this reaction forward, ATPase couples ATP synthesis during cellular respiration to an electrochemical gradient created by the difference in proton (H+) concentration across the mitochondrial membrane in eukaryotes or the plasma membrane in bacteria. During photosynthesis in plants, ATP is synthesized by ATPase using a proton gradient created in the thylakoid lumen through the thylakoid membrane and into the chloroplast stroma.
ATP synthase consists of two main subunits, FO and F1, which has a rotational motor mechanism allowing for ATP production.[1][2]
Nomenclature
The F1 fraction derives its name from the term "Fraction 1" and FO (written as a subscript letter "o", not "zero") derives its name from being the binding fraction for oligomycin, a type of naturally-derived antibiotic that is able to inhibit the FO unit of ATP synthase.[3][4] These functional regions consist of different protein subunits — refer to tables.
Structure and function
Located within the thylakoid membrane and the inner mitochondrial membrane, ATP synthase consists of two regions FO and F1. FO causes rotation of F1 and is made of c-ring and subunits a, b, d, F6. F1 is made of subunits. F1 has a water soluble par that can hydrolyze ATP. FO on the other hand has mainly hydrophobic regions. FO F1 creates a pathway for protons movement across the membrane.[7]
F1 region
The F1 portion of ATP synthase is hydrophilic and responsible for hydrolyzing ATP. Subunits and make a hexamer with 6 binding sites. Three of them are catalytically inactive and they bind ADP.
Other three subunits catalyze the ATP synthesis. The other F1 subunits are are a part of a rotational motor mechanism. subunit allows to go though conformational changes, i.e closed, half open and open states allows for ATP to be bound and released once synthesized. The F1 particle is large and can be seen in the transmission electron microscope by negative staining.[8] These are particles of 9 nm diameter that pepper the inner mitochondrial membrane.
| Subunit | Human Gene |
|---|---|
| alpha | ATP5A1, ATPAF2 |
| beta | ATP5B, ATPAF1, C16orf7 |
| gamma | ATP5C1 |
| delta | ATP5D |
| epsilon | ATP5E |
FO region

Fo is a water insoluble protein with eight subunits and transmembrane ring. The ring has a tetramer shape with protein helix loop helix that goes though conformational change when protonated and deprotonated allowing to push neighboring subunits to rotate causing the spinning of FO which then affects conformation of F1 also, resulting on switching of states of alpha and beta subunits. The FO region of ATP synthase is a proton pore that is embedded in the mitochondrial membrane. It consists of three main subunits a, b, and c, and (in humans) six additional subunits, d, e, f, g, F6, and 8 (or A6L).
| Subunit | Human Gene |
|---|---|
| a | ATP6 |
| b | ATP5F1 |
| c | ATP5G1, ATP5G2, ATP5G3 |
Binding model

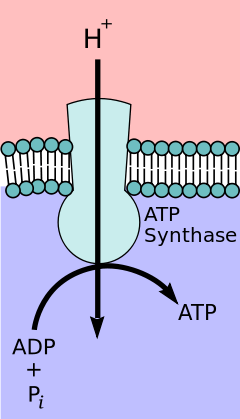
In the 1960s through the 1970s, Paul Boyer, a UCLA Professor, developed the binding change, or flip-flop, mechanism theory, which postulated that ATP synthesis is dependent on a conformational change in ATP synthase generated by rotation of the gamma subunit. The research group of John E. Walker, then at the MRC Laboratory of Molecular Biology in Cambridge, crystallized the F1 catalytic-domain of ATP synthase. The structure, at the time the largest asymmetric protein structure known, indicated that Boyer's rotary-catalysis model was, in essence, correct. For elucidating this, Boyer and Walker shared half of the 1997 Nobel Prize in Chemistry.
The crystal structure of the F1 showed alternating alpha and beta subunits (3 of each), arranged like segments of an orange around a rotating asymmetrical gamma subunit. According to the current model of ATP synthesis (known as the alternating catalytic model), the transmembrane potential created by (H+) proton cations supplied by the electron transport chain, drives the (H+) proton cations from the intermembrane space through the membrane via the FO region of ATP synthase. A portion of the FO (the ring of c-subunits) rotates as the protons pass through the membrane. The c-ring is tightly attached to the asymmetric central stalk (consisting primarily of the gamma subunit), causing it to rotate within the alpha3beta3 of F1 causing the 3 catalytic nucleotide binding sites to go through a series of conformational changes that leads to ATP synthesis. The major F1 subunits are prevented from rotating in sympathy with the central stalk rotor by a peripheral stalk that joins the alpha3beta3 to the non-rotating portion of FO. The structure of the intact ATP synthase is currently known at low-resolution from electron cryo-microscopy (cryo-EM) studies of the complex. The cryo-EM model of ATP synthase suggests that the peripheral stalk is a flexible structure that wraps around the complex as it joins F1 to FO. Under the right conditions, the enzyme reaction can also be carried out in reverse, with ATP hydrolysis driving proton pumping across the membrane.
The binding change mechanism involves the active site of a β subunit's cycling between three states.[10] In the "open" state, ADP and phosphate enter the active site; in the adjacent diagram, this is shown in red. The protein then closes up around the molecules and binds them loosely — the "loose" state (shown in orange). The enzyme then undergoes another change in shape and forces these molecules together, with the active site in the resulting "tight" state (shown in pink) binding the newly produced ATP molecule with very high affinity. Finally, the active site cycles back to the open state, releasing ATP and binding more ADP and phosphate, ready for the next cycle of ATP production.[11]
Physiological role
Like other enzymes, the activity of F1FO ATP synthase is reversible. Large-enough quantities of ATP cause it to create a transmembrane proton gradient, this is used by fermenting bacteria that do not have an electron transport chain, but rather hydrolyze ATP to make a proton gradient, which they use to drive flagella and the transport of nutrients into the cell.
In respiring bacteria under physiological conditions, ATP synthase, in general, runs in the opposite direction, creating ATP while using the proton motive force created by the electron transport chain as a source of energy. The overall process of creating energy in this fashion is termed oxidative phosphorylation. The same process takes place in the mitochondria, where ATP synthase is located in the inner mitochondrial membrane and the F1-part projects into mitochondrial matrix. The consumption of ATP by ATP-synthase pumps proton cations into the matrix.
Evolution
The evolution of ATP synthase is thought to have been modular whereby two functionally independent subunits became associated and gained new functionality.[12][13] This association appears to have occurred early in evolutionary history, because essentially the same structure and activity of ATP synthase enzymes are present in all kingdoms of life.[12] The F-ATP synthase displays high functional and mechanistic similarity to the V-ATPase.[14] However, whereas the F-ATP synthase generates ATP by utilising a proton gradient, the V-ATPase generates a proton gradient at the expense of ATP, generating pH values of as low as 1.[15]
The F1 region also shows significant similarity to hexameric DNA helicases, and the FO region shows some similarity to H+
-powered flagellar motor complexes.[14] The α3β3 hexamer of the F1 region shows significant structural similarity to hexameric DNA helicases; both form a ring with 3-fold rotational symmetry with a central pore. Both have roles dependent on the relative rotation of a macromolecule within the pore; the DNA helicases use the helical shape of DNA to drive their motion along the DNA molecule and to detect supercoiling, whereas the α3β3 hexamer uses the conformational changes through the rotation of the γ subunit to drive an enzymatic reaction.[16]
The H+
motor of the FO particle shows great functional similarity to the H+
motors that drive flagella.[14] Both feature a ring of many small alpha-helical proteins that rotate relative to nearby stationary proteins, using a H+
potential gradient as an energy source. This link is tenuous, however, as the overall structure of flagellar motors is far more complex than that of the FO particle and the ring with about 30 rotating proteins is far larger than the 10, 11, or 14 helical proteins in the FO complex.
The modular evolution theory for the origin of ATP synthase suggests that two subunits with independent function, a DNA helicase with ATPase activity and a H+
motor, were able to bind, and the rotation of the motor drove the ATPase activity of the helicase in reverse.[12][16] This complex then evolved greater efficiency and eventually developed into today's intricate ATP synthases. Alternatively, the DNA helicase/H+
motor complex may have had H+
pump activity with the ATPase activity of the helicase driving the H+
motor in reverse.[12] This may have evolved to carry out the reverse reaction and act as an ATP synthase.[13][17][18]
In different species
E. coli
E. coli ATP synthase is the simplest known form of ATP synthase, with 8 different subunit types.[19]
Yeast
Yeast ATP synthase is one of the best-studied eukaryotic ATP synthases; and five F1, eight FO subunits, and seven associated proteins have been identified.[7] Most of these proteins have homologues in other eukaryotes.[20][21][22]
Plant
In plants, ATP synthase is also present in chloroplasts (CF1FO-ATP synthase). The enzyme is integrated into thylakoid membrane; the CF1-part sticks into stroma, where dark reactions of photosynthesis (also called the light-independent reactions or the Calvin cycle) and ATP synthesis take place. The overall structure and the catalytic mechanism of the chloroplast ATP synthase are almost the same as those of the mitochondrial enzyme. However, in chloroplasts, the proton motive force is generated not by respiratory electron transport chain but by primary photosynthetic proteins.
Bovine
The ATP synthase isolated from bovine heart mitochondria (Bos taurus) is, in terms of biochemistry and structure, the best-characterized ATP synthase. Beef heart is used as a source for the enzyme because of the high concentration of mitochondria in cardiac muscle.[23][24][25]
Human
The following is a list of humans genes that encode components of ATP synthases:
- ATP5A1, ATP5AL1
- ATP5B, ATP5BL1
- ATP5C2, ATP5D, ATP5E, ATP5F1, ATP5G1, ATP5G2, ATP5G3, ATP5H, ATP5HP1, ATP5I, ATP5J, ATP5J2, ATP5L, ATP5L2, ATP5O, ATP5S
- ATP6, ATP6AP1, ATP6AP2
- ATPSBL1, ATPSBL2
- MT-ATP6, MT-ATP8
See also
- ATP10 protein required for the assembly of the FO sector of the mitochondrial ATPase complex.
- Chloroplast
- Electron transfer chain
- Flavoprotein
- Mitochondrion
- Oxidative phosphorylation
- P-ATPase
- Proton pump
- Rotating locomotion in living systems
- Transmembrane ATPase
- V-ATPase
References
- ↑ Okuno D, Iino R, Noji H (April 2011). "Rotation and structure of FOF1-ATP synthase". Journal of Biochemistry. 149 (6): 655–64. PMID 21524994. doi:10.1093/jb/mvr049.
- ↑ Junge W, Nelson N (June 2015). "ATP synthase". Annual Review of Biochemistry. 84: 631–57. PMID 25839341. doi:10.1146/annurev-biochem-060614-034124.
- ↑ Kagawa Y, Racker E (May 1966). "Partial resolution of the enzymes catalyzing oxidative phosphorylation. 8. Properties of a factor conferring oligomycin sensitivity on mitochondrial adenosine triphosphatase". The Journal of Biological Chemistry. 241 (10): 2461–6. PMID 4223640.
- ↑ Mccarty RE (November 1992). "A plant biochemist's view of H+
-ATPases and ATP synthases". The Journal of Experimental Biology. 172 (Pt 1): 431–441. PMID 9874753. - ↑ PDB: 5ARA; Zhou A, Rohou A, Schep DG, Bason JV, Montgomery MG, Walker JE, Grigorieff N, Rubinstein JL (October 2015). "Structure and conformational states of the bovine mitochondrial ATP synthase by cryo-EM". eLife. 4: e10180. PMC 4718723
 . PMID 26439008. doi:10.7554/eLife.10180.
. PMID 26439008. doi:10.7554/eLife.10180. - ↑ Goodsell D (December 2005). "ATP Synthase". Molecule of the Month. RCSB PDB. doi:10.2210/rcsb_pdb/mom_2005_12.
- 1 2 Velours J, Paumard P, Soubannier V, Spannagel C, Vaillier J, Arselin G, Graves PV (May 2000). "Organisation of the yeast ATP synthase F(0):a study based on cysteine mutants, thiol modification and cross-linking reagents". Biochimica et Biophysica Acta. 1458 (2–3): 443–56. PMID 10838057. doi:10.1016/S0005-2728(00)00093-1.
- ↑ Fernandez Moran H, Oda T, Blair PV, Green DE (July 1964). "A macromolecular repeating unit of mitochondrial structure and function. Correlated electron microscopic and biochemical studies of isolated mitochondria and submitochondrial particles of beef heart muscle". The Journal of Cell Biology. 22 (1): 63–100. PMC 2106494
 . PMID 14195622. doi:10.1083/jcb.22.1.63.
. PMID 14195622. doi:10.1083/jcb.22.1.63. - ↑ PDB: 1VZS; Carbajo RJ, Silvester JA, Runswick MJ, Walker JE, Neuhaus D (2004). "Solution structure of subunit F(6) from the peripheral stalk region of ATP synthase from bovine heart mitochondria". Journal of Molecular Biology. 342 (2): 593–603. PMID 15327958. doi:10.1016/j.jmb.2004.07.013.
- ↑ Gresser MJ, Myers JA, Boyer PD (October 1982). "Catalytic site cooperativity of beef heart mitochondrial F1 adenosine triphosphatase. Correlations of initial velocity, bound intermediate, and oxygen exchange measurements with an alternating three-site model". The Journal of Biological Chemistry. 257 (20): 12030–8. PMID 6214554.
- ↑ Nakamoto RK, Baylis Scanlon JA, Al-Shawi MK (August 2008). "The rotary mechanism of the ATP synthase". Archives of Biochemistry and Biophysics. 476 (1): 43–50. PMC 2581510
 . PMID 18515057. doi:10.1016/j.abb.2008.05.004.
. PMID 18515057. doi:10.1016/j.abb.2008.05.004. - 1 2 3 4 Doering C, Ermentrout B, Oster G (December 1995). "Rotary DNA motors". Biophysical Journal. 69 (6): 2256–67. Bibcode:1995BpJ....69.2256D. PMC 1236464
 . PMID 8599633. doi:10.1016/S0006-3495(95)80096-2.
. PMID 8599633. doi:10.1016/S0006-3495(95)80096-2. - 1 2 Crofts A. "Lecture 10:ATP synthase". Life Sciences at the University of Illinois at Urbana-Champaign.
- 1 2 3 "ATP Synthase". InterPro Database.
- ↑ Kim MS, Jang J, Ab Rahman NB, Pethe K, Berry EA, Huang LS (June 2015). "Isolation and Characterization of a Hybrid Respiratory Supercomplex Consisting of Mycobacterium tuberculosis Cytochrome bcc and Mycobacterium smegmatis Cytochrome aa3". The Journal of Biological Chemistry. 290 (23): 14350–60. PMC 4505504
 . PMID 25861988. doi:10.1074/jbc.m114.624312.
. PMID 25861988. doi:10.1074/jbc.m114.624312. - 1 2 Martinez LO, Jacquet S, Esteve JP, Rolland C, Cabezón E, Champagne E, Pineau T, Georgeaud V, Walker JE, Tercé F, Collet X, Perret B, Barbaras R (January 2003). "Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis". Nature. 421 (6918): 75–9. Bibcode:2003Natur.421...75M. PMID 12511957. doi:10.1038/nature01250.
- ↑ Cross RL, Taiz L (January 1990). "Gene duplication as a means for altering H+/ATP ratios during the evolution of FOF1 ATPases and synthases". FEBS Letters. 259 (2): 227–9. PMID 2136729. doi:10.1016/0014-5793(90)80014-a.
- ↑ Cross RL, Müller V (October 2004). "The evolution of A-, F-, and V-type ATP synthases and ATPases: reversals in function and changes in the H+/ATP coupling ratio". FEBS Letters. 576 (1–2): 1–4. PMID 15473999. doi:10.1016/j.febslet.2004.08.065.
- ↑ Ahmad Z, Okafor F, Laughlin TF. "Role of Charged Residues in the Catalytic Sites of Escherichia coli ATP Synthase". Journal of Amino Acids. 2011: 785741. PMID 22312470. doi:10.4061/2011/785741.
- ↑ Devenish RJ, Prescott M, Roucou X, Nagley P (May 2000). "Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex". Biochimica et Biophysica Acta. 1458 (2–3): 428–42. PMID 10838056. doi:10.1016/S0005-2728(00)00092-X.
- ↑ Kabaleeswaran V, Puri N, Walker JE, Leslie AG, Mueller DM (November 2006). "Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase". The EMBO Journal. 25 (22): 5433–42. PMID 17082766. doi:10.1038/sj.emboj.7601410.
- ↑ Stock D, Leslie AG, Walker JE (November 1999). "Molecular architecture of the rotary motor in ATP synthase". Science. 286 (5445): 1700–5. PMID 10576729.
- ↑ Abrahams JP, Leslie AG, Lutter R, Walker JE (August 1994). "Structure at 2.8 A resolution of F1-ATPase from bovine heart mitochondria". Nature. 370 (6491): 621–8. PMID 8065448. doi:10.1038/370621a0.
- ↑ Gibbons C, Montgomery MG, Leslie AG, Walker JE (November 2000). "The structure of the central stalk in bovine F(1)-ATPase at 2.4 A resolution". Nature Structural Biology. 7 (11): 1055–61. PMID 11062563. doi:10.1038/80981.
- ↑ Menz RI, Walker JE, Leslie AG (August 2001). "Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: implications for the mechanism of rotary catalysis". Cell. 106 (3): 331–41. PMID 11509182.
External links
- "ATP synthase — a splendid molecular machine"
- Well illustrated ATP synthase lecture by Antony Crofts of the University of Illinois at Urbana-Champaign.
- Proton and Sodium translocating F-type, V-type and A-type ATPases in OPM database
- The Nobel Prize in Chemistry 1997 to Paul D. Boyer and John E. Walker for the enzymatic mechanism of synthesis of ATP; and to Jens C. Skou, for discovery of an ion-transporting enzyme, Na+
, K+
-ATPase. - Harvard Multimedia Production Site — Videos — ATP synthesis animation
- "ATP Synthase- Molecule of the Month"
