Apolipoprotein E
Apolipoprotein E (ApoE) is a class of apolipoprotein found in the chylomicron and Intermediate-density lipoprotein (IDLs) that is essential for the normal catabolism of triglyceride-rich lipoprotein constituents.[3] In peripheral tissues, ApoE is primarily produced by the liver and macrophages, and mediates cholesterol metabolism in an isoform-dependent manner. In the central nervous system, ApoE is mainly produced by astrocytes, and transports cholesterol to neurons via ApoE receptors, which are members of the low density lipoprotein receptor gene family.[4] ApoE is the principal cholesterol carrier in the brain.[5] This protein is involved in Alzheimer's disease and cardiovascular disease.[6]
Structure
Gene
The gene, APOE, is mapped to chromosome 19 in a cluster with apolipoprotein C1 and the apolipoprotein C2. The APOE gene consists of four exons and three introns, totaling 3597 base pairs. APOE is transcriptionally activated by the liver X receptor (an important regulator of cholesterol, fatty acid, and glucose homeostasis) and peroxisome proliferator-activated receptor γ, nuclear receptors that form heterodimers with retinoid X receptors.[7] In melanocytic cells APOE gene expression may be regulated by MITF.[8]
Protein
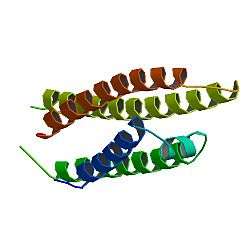
APOE is 299 amino acids long and contains multiple amphipathic α-helices. According to crystallography studies, a hinge region connects the N- and C-terminal regions of the protein. The N-terminal region (residues 1–167) forms an anti-parallel four-helix bundle such that the non-polar sides face inside the protein. Meanwhile, the C-terminal domain (residues 206-299) contains three α-helices which form a large exposed hydrophobic surface and interact with those in the N-terminal helix bundle domain through hydrogen bonds and salt-bridges. The C-terminal region also contains a low density lipoprotein receptor (LDLR)-binding site.[9]
Polymorphisms
APOE is polymorphic,[10][11] with three major alleles: APOE-ε2 (cys112, cys158), APOE-ε3 (cys112, arg158), and APOE-ε4 (arg112, arg158).[6][12][13] Although these allelic forms differ from each other by only one or two amino acids at positions 112 and 158,[14][15][16] these differences alter APOE structure and function. These have physiological consequences:
- ε2 (rs7412-T, rs429358-T) has an allele frequency of approximately 7 percent.[17] This variant of the apoprotein binds poorly to cell surface receptors while E3 and E4 bind well.[18] E2 is associated with both increased and decreased risk for atherosclerosis. Individuals with an E2/E2 combination may clear dietary fat slowly and be at greater risk for early vascular disease and the genetic disorder type III hyperlipoproteinemia—94.4% of such patients are E2/E2, while only ∼2% of E2/E2 develop the disease, so other environmental and genetic factors are likely to be involved (such as cholesterol in the diet and age).[19][20][21] E2 has also been implicated in Parkinson's disease,[22] but this finding was not replicated in a larger population association study.[23]
- ε3 (rs7412-C, rs429358-T) has an allele frequency of approximately 79 percent.[17] It is considered the "neutral" Apo E genotype.
- ε4 (rs7412-C, rs429358-C) has an allele frequency of approximately 14 percent.[17] E4 has been implicated in atherosclerosis,[24] Alzheimer's disease,[25][26] impaired cognitive function,[27][28] reduced hippocampal volume,[28] HIV,[29] faster disease progression in multiple sclerosis,[30][31] unfavorable outcome after traumatic brain injury,[32] ischemic cerebrovascular disease,[33] sleep apnea,[34][35] accelerated telomere shortening [36] and reduced neurite outgrowth.[37] A notable advantage of the E4 allele (relative to E2 and E3) is a positive association with higher levels of vitamin D, which may help explain its prevalence despite its seeming complicity in various diseases or disorders.[38]
However, there is much to be learned about these APOE isoforms, including the interaction of other potentially protective genetic polymorphisms, so caution is advised before making determinant statements about the influence of APOE polymorphisms; this is particularly true as it relates to how APOE isoforms influence cognition and the development of Alzheimer’s Disease. In addition, there is no evidence that APOE polymorphisms influence cognition in younger age groups (other than possible increased episodic memory ability and neural efficiency in younger APOE4 age groups), nor is there evidence that the APOE4 isoform places individuals at increased risk for any infectious disease.[39]
Function
APOE transports lipoproteins, fat-soluble vitamins, and cholesterol into the lymph system and then into the blood. It is synthesized principally in the liver, but has also been found in other tissues such as the brain, kidneys, and spleen.[12] In the nervous system, non-neuronal cell types, most notably astroglia and microglia, are the primary producers of APOE, while neurons preferentially express the receptors for APOE.[40] There are seven currently identified mammalian receptors for APOE which belong to the evolutionarily conserved LDLR family.[41]
APOE was initially recognized for its importance in lipoprotein metabolism and cardiovascular disease. Defects in APOE result in familial dysbetalipoproteinemia aka type III hyperlipoproteinemia (HLP III), in which increased plasma cholesterol and triglycerides are the consequence of impaired clearance of chylomicron, VLDL and LDL remnants.[3] More recently, it has been studied for its role in several biological processes not directly related to lipoprotein transport, including Alzheimer's disease (AD), immunoregulation, and cognition.[6] Though the exact mechanisms remain to be elucidated, isoform 4 of APOE, encoded by an APOE allele, has been associated with increased calcium ion levels and apoptosis following mechanical injury.[42]
In the field of immune regulation, a growing number of studies point to APOE's interaction with many immunological processes, including suppressing T cell proliferation, macrophage functioning regulation, lipid antigen presentation facilitation (by CD1) [43] to natural killer T cell as well as modulation of inflammation and oxidation.[44] APOE is produced by macrophages and APOE secretion has been shown to be restricted to classical monocytes in PBMC, and the secretion of APOE by monocytes is down regulated by inflammatory cytokines and upregulated by TGF-beta.[45]
Clinical significance
Alzheimer's disease
The E4 variant is the largest known genetic risk factor for late-onset sporadic Alzheimer's disease (AD) in a variety of ethnic groups.[46] "Nigerian blacks have the highest observed frequency of the APO E*4 allele in world populations."[47] But AD is rare among them.[47][48] There is growing evidence that suggests that this may be due to their low cholesterol levels.[47][48][49][50] Caucasian and Japanese carriers of 2 E4 alleles have between 10 and 30 times the risk of developing AD by 75 years of age, as compared to those not carrying any E4 alleles. While the exact mechanism of how E4 causes such dramatic effects remains to be fully determined, evidence has been presented suggesting an interaction with amyloid.[51] Alzheimer's disease is characterized by build-ups of aggregates of the peptide beta-amyloid. Apolipoprotein E enhances proteolytic break-down of this peptide, both within and between cells. The isoform ApoE-ε4 is not as effective as the others at promoting these reactions, resulting in increased vulnerability to AD in individuals with that gene variation.[52]
Although 40-65% of AD patients have at least one copy of the ε4 allele, ApoE4 is not a determinant of the disease - at least a third of patients with AD are ApoE4 negative and some ApoE4 homozygotes never develop the disease. Yet those with two ε4 alleles have up to 20 times the risk of developing AD.[53] There is also evidence that the ApoE2 allele may serve a protective role in AD.[54] Thus, the genotype most at risk for Alzheimer's disease and at an earlier age is ApoE 4,4. Using genotype ApoE 3,3 as a benchmark (with the persons who have this genotype regarded as having a risk level of 1.0), individuals with genotype ApoE4,4 have an odds ratio of 14.9 of developing Alzheimer's disease. Individuals with the ApoE 3,4 genotype face an odds ratio of 3.2, and people with a copy of the 2 allele and the 4 allele (ApoE2,4), have an odds ratio of 2.6. Persons with one copy each of the 2 allele and the 3 allele (ApoE2,3) have an odds ratio of 0.6. Persons with two copies of the 2 allele (ApoE2,2) also have an odds ratio of 0.6.[55]
| Estimated worldwide human allele frequencies of ApoE * in Caucasian population[55] | ||||
| Allele | ε2 | ε3 | ε4 | |
|---|---|---|---|---|
| General Frequency | 8.4% | 77.9% | 13.7% | |
| AD Frequency | 3.9% | 59.4% | 36.7% | |
While ApoE4 has been found to greatly increase the odds that an individual will develop Alzheimer’s, a 2002 study concluded, that in persons with any combination of ApoE alleles, high serum total cholesterol and high blood pressure in mid-life are independent risk factors which together can nearly triple the risk that the individual will later develop AD.[50] Projecting from their data, some researchers have suggested that lowering serum cholesterol levels may reduce a person’s risk for Alzheimer’s disease, even if they have two ApoE4 alleles, thus reducing the risk from nine or ten times the odds of getting AD down to just two times the odds.[50]
Atherosclerosis
Knockout mice that lack the apolipoprotein-E gene (ApoE−/−) develop extreme hypercholesterolemia when fed a high-fat diet.[56]
Interactions
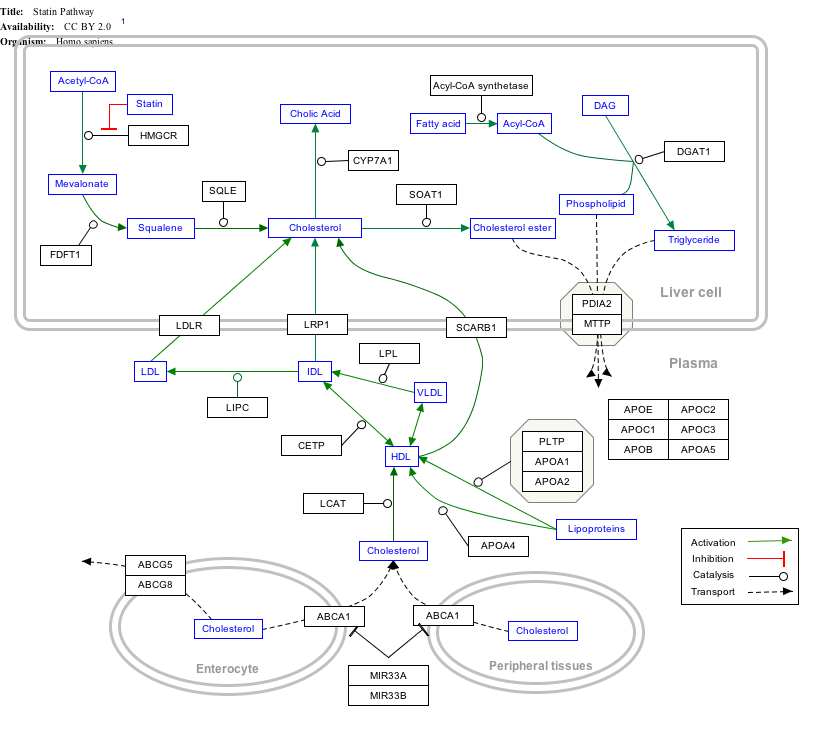
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
|px|alt=Statin Pathway edit]]
Statin Pathway edit
- ↑ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
References
- ↑ "Human PubMed Reference:".
- ↑ "Mouse PubMed Reference:".
- 1 2 "Entrez Gene: APOE apolipoprotein E".
- ↑ Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G (February 2013). "Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy". Nature Reviews. Neurology. 9 (2): 106–18. PMC 3726719
 . PMID 23296339. doi:10.1038/nrneurol.2012.263.
. PMID 23296339. doi:10.1038/nrneurol.2012.263. - ↑ Puglielli L, Tanzi RE, Kovacs DM (April 2003). "Alzheimer's disease: the cholesterol connection". Nature Neuroscience. 6 (4): 345–51. PMID 12658281. doi:10.1038/nn0403-345.
- 1 2 3 (ed.), Ian P. Stolerman (2010). Encyclopedia of psychopharmacology (Online-Ausg. ed.). Berlin: Springer. ISBN 9783540686989.
- ↑ Chawla A, Boisvert WA, Lee CH, Laffitte BA, Barak Y, Joseph SB, Liao D, Nagy L, Edwards PA, Curtiss LK, Evans RM, Tontonoz P (January 2001). "A PPAR gamma-LXR-ABCA1 pathway in macrophages is involved in cholesterol efflux and atherogenesis". Molecular Cell. 7 (1): 161–71. PMID 11172721. doi:10.1016/S1097-2765(01)00164-2.
- ↑ Hoek KS, Schlegel NC, Eichhoff OM, Widmer DS, Praetorius C, Einarsson SO, Valgeirsdottir S, Bergsteinsdottir K, Schepsky A, Dummer R, Steingrimsson E (December 2008). "Novel MITF targets identified using a two-step DNA microarray strategy". Pigment Cell & Melanoma Research. 21 (6): 665–76. PMID 19067971. doi:10.1111/j.1755-148X.2008.00505.x.
- ↑ Phillips MC (September 2014). "Apolipoprotein E isoforms and lipoprotein metabolism". IUBMB Life. 66 (9): 616–23. PMID 25328986. doi:10.1002/iub.1314.
- ↑ Singh PP, Singh M, Mastana SS (2006). "APOE distribution in world populations with new data from India and the UK". Annals of Human Biology. 33 (3): 279–308. PMID 17092867. doi:10.1080/03014460600594513.
- ↑ Eisenberg DT, Kuzawa CW, Hayes MG (September 2010). "Worldwide allele frequencies of the human apolipoprotein E gene: climate, local adaptations, and evolutionary history". American Journal of Physical Anthropology. 143 (1): 100–11. PMID 20734437. doi:10.1002/ajpa.21298.
- 1 2 Baars H, van der Smagt J, Doevandans P (2011). Clinical Cardiogenetics. London: Springer. ISBN 9781849964715.
- ↑ Ghebranious N, Ivacic L, Mallum J, Dokken C (2005). "Detection of ApoE E2, E3 and E4 alleles using MALDI-TOF mass spectrometry and the homogeneous mass-extend technology". Nucleic Acids Research. 33 (17): e149. PMC 1243648
 . PMID 16204452. doi:10.1093/nar/gni155.
. PMID 16204452. doi:10.1093/nar/gni155. - ↑ Online Mendelian Inheritance in Man (OMIM) APOE3 isoform, hyperlipoproteinemia, type III, autosomal recessive -107741#0015
- ↑ Online Mendelian Inheritance in Man (OMIM) APOE3 isoform, APOE, CYS112 and ARG158 -107741#0001
- ↑ Zuo L, van Dyck CH, Luo X, Kranzler HR, Yang BZ, Gelernter J (2006). "Variation at APOE and STH loci and Alzheimer's disease". Behavioral and Brain Functions. 2 (1): 13. PMC 1526745
 . PMID 16603077. doi:10.1186/1744-9081-2-13.
. PMID 16603077. doi:10.1186/1744-9081-2-13. - 1 2 3 "Alzheimer Research Forum: Meta-Analyses of apolipoprotein E AD Association Studies". Archived from the original on 2012-11-24.
- ↑ Weisgraber KH, Innerarity TL, Mahley RW (March 1982). "Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site". The Journal of Biological Chemistry. 257 (5): 2518–21. PMID 6277903.
- ↑ Breslow JL, Zannis VI, SanGiacomo TR, Third JL, Tracy T, Glueck CJ (November 1982). "Studies of familial type III hyperlipoproteinemia using as a genetic marker the apoE phenotype E2/2". Journal of Lipid Research. 23 (8): 1224–35. PMID 7175379.
- ↑ Feussner G, Feussner V, Hoffmann MM, Lohrmann J, Wieland H, März W (1998). "Molecular basis of type III hyperlipoproteinemia in Germany". Human Mutation. 11 (6): 417–23. PMID 9603433. doi:10.1002/(SICI)1098-1004(1998)11:6<417::AID-HUMU1>3.0.CO;2-5.
- ↑ Civeira F, Pocoví M, Cenarro A, Casao E, Vilella E, Joven J, González J, Garcia-Otín AL, Ordovás JM (December 1996). "Apo E variants in patients with type III hyperlipoproteinemia". Atherosclerosis. 127 (2): 273–82. PMID 9125318. doi:10.1016/S0021-9150(96)05969-2.
- ↑ Huang X, Chen PC, Poole C (June 2004). "APOE-[epsilon]2 allele associated with higher prevalence of sporadic Parkinson disease". Neurology. 62 (12): 2198–202. PMID 15210882. doi:10.1212/01.wnl.0000130159.28215.6a.
- ↑ Federoff M, Jimenez-Rolando B, Nalls MA, Singleton AB (May 2012). "A large study reveals no association between APOE and Parkinson's disease". Neurobiology of Disease. 46 (2): 389–92. PMC 3323723
 . PMID 22349451. doi:10.1016/j.nbd.2012.02.002.
. PMID 22349451. doi:10.1016/j.nbd.2012.02.002. - ↑ Mahley RW (April 1988). "Apolipoprotein E: cholesterol transport protein with expanding role in cell biology". Science. 240 (4852): 622–30. PMID 3283935. doi:10.1126/science.3283935.
- ↑ Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA (August 1993). "Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families". Science. 261 (5123): 921–3. PMID 8346443. doi:10.1126/science.8346443.
- ↑ Strittmatter WJ, Saunders AM, Schmechel D, Pericak-Vance M, Enghild J, Salvesen GS, Roses AD (March 1993). "Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease". Proceedings of the National Academy of Sciences of the United States of America. 90 (5): 1977–81. PMC 46003
 . PMID 8446617. doi:10.1073/pnas.90.5.1977.
. PMID 8446617. doi:10.1073/pnas.90.5.1977. - ↑ Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF, Carothers A, Whalley LJ (August 2002). "Cognitive change and the APOE epsilon 4 allele". Nature. 418 (6901): 932. PMID 12198535. doi:10.1038/418932a.
- 1 2 Farlow MR, He Y, Tekin S, Xu J, Lane R, Charles HC (November 2004). "Impact of APOE in mild cognitive impairment". Neurology. 63 (10): 1898–901. PMID 15557508. doi:10.1212/01.wnl.0000144279.21502.b7.
- ↑ Burt TD, Agan BK, Marconi VC, He W, Kulkarni H, Mold JE, Cavrois M, Huang Y, Mahley RW, Dolan MJ, McCune JM, Ahuja SK (June 2008). "Apolipoprotein (apo) E4 enhances HIV-1 cell entry in vitro, and the APOE epsilon4/epsilon4 genotype accelerates HIV disease progression". Proceedings of the National Academy of Sciences of the United States of America. 105 (25): 8718–23. PMC 2438419
 . PMID 18562290. doi:10.1073/pnas.0803526105.
. PMID 18562290. doi:10.1073/pnas.0803526105. - ↑ Chapman J, Vinokurov S, Achiron A, Karussis DM, Mitosek-Szewczyk K, Birnbaum M, Michaelson DM, Korczyn AD (February 2001). "APOE genotype is a major predictor of long-term progression of disability in MS". Neurology. 56 (3): 312–6. PMID 11171894. doi:10.1212/wnl.56.3.312.
- ↑ Schmidt S, Barcellos LF, DeSombre K, Rimmler JB, Lincoln RR, Bucher P, Saunders AM, Lai E, Martin ER, Vance JM, Oksenberg JR, Hauser SL, Pericak-Vance MA, Haines JL (March 2002). "Association of polymorphisms in the apolipoprotein E region with susceptibility to and progression of multiple sclerosis". American Journal of Human Genetics. 70 (3): 708–17. PMC 384947
 . PMID 11836653. doi:10.1086/339269.
. PMID 11836653. doi:10.1086/339269. - ↑ Friedman G, Froom P, Sazbon L, Grinblatt I, Shochina M, Tsenter J, Babaey S, Yehuda B, Groswasser Z (January 1999). "Apolipoprotein E-epsilon4 genotype predicts a poor outcome in survivors of traumatic brain injury". Neurology. 52 (2): 244–8. PMID 9932938. doi:10.1212/wnl.52.2.244.
- ↑ McCarron MO, Delong D, Alberts MJ (October 1999). "APOE genotype as a risk factor for ischemic cerebrovascular disease: a meta-analysis". Neurology. 53 (6): 1308–11. PMID 10522889. doi:10.1212/wnl.53.6.1308.
- ↑ Kadotani H, Kadotani T, Young T, Peppard PE, Finn L, Colrain IM, Murphy GM, Mignot E (June 2001). "Association between apolipoprotein E epsilon4 and sleep-disordered breathing in adults". JAMA. 285 (22): 2888–90. PMID 11401610. doi:10.1001/jama.285.22.2888.
- ↑ Gottlieb DJ, DeStefano AL, Foley DJ, Mignot E, Redline S, Givelber RJ, Young T (August 2004). "APOE epsilon4 is associated with obstructive sleep apnea/hypopnea: the Sleep Heart Health Study". Neurology. 63 (4): 664–8. PMID 15326239. doi:10.1212/01.wnl.0000134671.99649.32.
- ↑ Jacobs EG, Kroenke C, Lin J, Epel ES, Kenna HA, Blackburn EH, Rasgon NL (February 2013). "Accelerated cell aging in female APOE-ε4 carriers: implications for hormone therapy use". PloS One. 8 (2): e54713. PMC 3572118
 . PMID 23418430. doi:10.1371/journal.pone.0054713.
. PMID 23418430. doi:10.1371/journal.pone.0054713. - ↑ Raber J (May 2008). "AR, apoE, and cognitive function". Hormones and Behavior. 53 (5): 706–15. PMC 2409114
 . PMID 18395206. doi:10.1016/j.yhbeh.2008.02.012.
. PMID 18395206. doi:10.1016/j.yhbeh.2008.02.012. - ↑ Huebbe P, Nebel A, Siegert S, Moehring J, Boesch-Saadatmandi C, Most E, Pallauf J, Egert S, Müller MJ, Schreiber S, Nöthlings U, Rimbach G (September 2011). "APOE ε4 is associated with higher vitamin D levels in targeted replacement mice and humans". FASEB Journal. 25 (9): 3262–70. PMID 21659554. doi:10.1096/fj.11-180935.
- ↑ Mondadori CR, de Quervain DJ, Buchmann A, Mustovic H, Wollmer MA, Schmidt CF, Boesiger P, Hock C, Nitsch RM, Papassotiropoulos A, Henke K (August 2007). "Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers". Cerebral Cortex. 17 (8): 1934–47. PMID 17077159. doi:10.1093/cercor/bhl103.
- ↑ Zhang Z, Mu J, Li J, Li W, Song J (January 2013). "Aberrant apolipoprotein E expression and cognitive dysfunction in patients with poststroke depression". Genetic Testing and Molecular Biomarkers. 17 (1): 47–51. PMC 3525887
 . PMID 23171142. doi:10.1089/gtmb.2012.0253.
. PMID 23171142. doi:10.1089/gtmb.2012.0253. - ↑ Rogers JT, Weeber EJ (August 2008). "Reelin and apoE actions on signal transduction, synaptic function and memory formation". Neuron Glia Biology. 4 (3): 259–70. PMID 19674510. doi:10.1017/S1740925X09990184.
- ↑ Jiang L, Zhong J, Dou X, Cheng C, Huang Z, Sun X (Aug 2015). "Effects of ApoE on intracellular calcium levels and apoptosis of neurons after mechanical injury". Neuroscience. 301: 375–83. PMID 26073697. doi:10.1016/j.neuroscience.2015.06.005.
- ↑ van den Elzen P, Garg S, León L, Brigl M, Leadbetter EA, Gumperz JE, Dascher CC, Cheng TY, Sacks FM, Illarionov PA, Besra GS, Kent SC, Moody DB, Brenner MB (October 2005). "Apolipoprotein-mediated pathways of lipid antigen presentation". Nature. 437 (7060): 906–10. PMID 16208376. doi:10.1038/nature04001.
- ↑ Zhang HL, Wu J, Zhu J (2010). "The role of apolipoprotein E in Guillain–Barré syndrome and experimental autoimmune neuritis". Journal of Biomedicine & Biotechnology. 2010: 357412. PMC 2825561
 . PMID 20182542. doi:10.1155/2010/357412.
. PMID 20182542. doi:10.1155/2010/357412. - ↑ Braesch-Andersen S, Paulie S, Smedman C, Mia S, Kumagai-Braesch M (2013). "ApoE production in human monocytes and its regulation by inflammatory cytokines". PloS One. 8 (11): e79908. PMC 3828220
 . PMID 24244577. doi:10.1371/journal.pone.0079908.
. PMID 24244577. doi:10.1371/journal.pone.0079908. - ↑ Sadigh-Eteghad S, Talebi M, Farhoudi M (October 2012). "Association of apolipoprotein E epsilon 4 allele with sporadic late onset Alzheimer`s disease. A meta-analysis". Neurosciences. 17 (4): 321–6. PMID 23022896.
- 1 2 3 Sepehrnia B, Kamboh MI, Adams-Campbell LL, Bunker CH, Nwankwo M, Majumder PP, Ferrell RE (October 1989). "Genetic studies of human apolipoproteins. X. The effect of the apolipoprotein E polymorphism on quantitative levels of lipoproteins in Nigerian blacks". American Journal of Human Genetics. 45 (4): 586–91. PMC 1683508
 . PMID 2491016.
. PMID 2491016. - 1 2 Notkola IL, Sulkava R, Pekkanen J, Erkinjuntti T, Ehnholm C, Kivinen P, Tuomilehto J, Nissinen A (1998-01-01). "Serum total cholesterol, apolipoprotein E epsilon 4 allele, and Alzheimer's disease". Neuroepidemiology. 17 (1): 14–20. PMID 9549720. doi:10.1159/000026149.
- ↑ Petanceska SS, DeRosa S, Sharma A, Diaz N, Duff K, Tint SG, Refolo LM, Pappolla M (2003-01-01). "Changes in apolipoprotein E expression in response to dietary and pharmacological modulation of cholesterol". Journal of Molecular Neuroscience. 20 (3): 395–406. PMID 14501024. doi:10.1385/JMN:20:3:395.
- 1 2 3 Kivipelto M, Helkala EL, Laakso MP, Hänninen T, Hallikainen M, Alhainen K, Iivonen S, Mannermaa A, Tuomilehto J, Nissinen A, Soininen H (August 2002). "Apolipoprotein E epsilon4 allele, elevated midlife total cholesterol level, and high midlife systolic blood pressure are independent risk factors for late-life Alzheimer disease". Annals of Internal Medicine. 137 (3): 149–55. PMID 12160362. doi:10.7326/0003-4819-137-3-200208060-00006.
- ↑ Wisniewski T, Frangione B (February 1992). "Apolipoprotein E: a pathological chaperone protein in patients with cerebral and systemic amyloid". Neuroscience Letters. 135 (2): 235–8. PMID 1625800. doi:10.1016/0304-3940(92)90444-C.
- ↑ Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, Mann K, Lamb B, Willson TM, Collins JL, Richardson JC, Smith JD, Comery TA, Riddell D, Holtzman DM, Tontonoz P, Landreth GE (June 2008). "ApoE promotes the proteolytic degradation of Abeta". Neuron. Cell Press. 58 (5): 681–93. PMC 2493297
 . PMID 18549781. doi:10.1016/j.neuron.2008.04.010. Lay summary – ScienceDaily (2008-06-13).
. PMID 18549781. doi:10.1016/j.neuron.2008.04.010. Lay summary – ScienceDaily (2008-06-13). - ↑ Hauser PS, Ryan RO (October 2013). "Impact of apolipoprotein E on Alzheimer's disease". Current Alzheimer Research. 10 (8): 809–17. PMC 3995977
 . PMID 23919769. doi:10.2174/15672050113109990156.
. PMID 23919769. doi:10.2174/15672050113109990156. - ↑ Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Rimmler JB, Locke PA, Conneally PM, Schmader KE (June 1994). "Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease". Nature Genetics. 7 (2): 180–4. PMID 7920638. doi:10.1038/ng0694-180.
- 1 2 Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM (1997). "Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. APOE and Alzheimer Disease Meta Analysis Consortium". JAMA. 278 (16): 1349–56. PMID 9343467. doi:10.1001/jama.1997.03550160069041.
- ↑ McNeill E, Channon KM, Greaves DR (June 2010). "Inflammatory cell recruitment in cardiovascular disease: murine models and potential clinical applications". Clinical Science. 118 (11): 641–55. PMID 20210786. doi:10.1042/CS20090488.
Further reading
- Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G (February 2013). "Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy". Nature Reviews. Neurology. 9 (2): 106–18. PMC 3726719
 . PMID 23296339. doi:10.1038/nrneurol.2012.263.
. PMID 23296339. doi:10.1038/nrneurol.2012.263. - Gunzburg MJ, Perugini MA, Howlett GJ (December 2007). "Structural basis for the recognition and cross-linking of amyloid fibrils by human apolipoprotein E". The Journal of Biological Chemistry. 282 (49): 35831–41. PMID 17916554. doi:10.1074/jbc.M706425200.
- Kolovou GD, Anagnostopoulou KK (August 2007). "Apolipoprotein E polymorphism, age and coronary heart disease". Ageing Research Reviews. 6 (2): 94–108. PMID 17224309. doi:10.1016/j.arr.2006.11.001.
- Lambert JC, Amouyel P (August 2007). "Genetic heterogeneity of Alzheimer's disease: complexity and advances". Psychoneuroendocrinology. 32 Suppl 1: S62–70. PMID 17659844. doi:10.1016/j.psyneuen.2007.05.015.
- Raber J (2007). "Role of apolipoprotein E in anxiety". Neural Plasticity. 2007: 91236. PMC 1940061
 . PMID 17710250. doi:10.1155/2007/91236.
. PMID 17710250. doi:10.1155/2007/91236. - Ye J (August 2007). "Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus". PLoS Pathogens. 3 (8): e108. PMC 1959368
 . PMID 17784784. doi:10.1371/journal.ppat.0030108.
. PMID 17784784. doi:10.1371/journal.ppat.0030108. - Bennet AM, Di Angelantonio E, Ye Z, Wensley F, Dahlin A, Ahlbom A, Keavney B, Collins R, Wiman B, de Faire U, Danesh J (September 2007). "Association of apolipoprotein E genotypes with lipid levels and coronary risk". JAMA. 298 (11): 1300–11. PMID 17878422. doi:10.1001/jama.298.11.1300.
- Itzhaki RF, Dobson CB, Shipley SJ, Wozniak MA (June 2004). "The role of viruses and of APOE in dementia". Annals of the New York Academy of Sciences. 1019 (1): 15–8. PMID 15246985. doi:10.1196/annals.1297.003.
- Ashford JW (2004). "APOE genotype effects on Alzheimer's disease onset and epidemiology". Journal of Molecular Neuroscience. 23 (3): 157–65. PMID 15181244. doi:10.1385/JMN:23:3:157.
- Huang Y, Weisgraber KH, Mucke L, Mahley RW (2004). "Apolipoprotein E: diversity of cellular origins, structural and biophysical properties, and effects in Alzheimer's disease". Journal of Molecular Neuroscience. 23 (3): 189–204. PMID 15181247. doi:10.1385/JMN:23:3:189.
- Masterman T, Hillert J (June 2004). "The telltale scan: APOE epsilon4 in multiple sclerosis". The Lancet. Neurology. 3 (6): 331. PMID 15157846. doi:10.1016/S1474-4422(04)00763-X.
- Bocksch L, Stephens T, Lucas A, Singh B (December 2001). "Apolipoprotein E: possible therapeutic target for atherosclerosis". Current Drug Targets. Cardiovascular & Haematological Disorders. 1 (2): 93–106. PMID 12769659. doi:10.2174/1568006013337944.
- Mahley RW, Rall SC (2002). "Apolipoprotein E: far more than a lipid transport protein". Annual Review of Genomics and Human Genetics. 1 (1): 507–37. PMID 11701639. doi:10.1146/annurev.genom.1.1.507.
- Parasuraman R, Greenwood PM, Sunderland T (April 2002). "The apolipoprotein E gene, attention, and brain function". Neuropsychology. 16 (2): 254–74. PMC 1350934
 . PMID 11949718. doi:10.1037/0894-4105.16.2.254.
. PMID 11949718. doi:10.1037/0894-4105.16.2.254. - Mahley RW, Ji ZS (January 1999). "Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E". Journal of Lipid Research. 40 (1): 1–16. PMID 9869645.
- Beffert U, Danik M, Krzywkowski P, Ramassamy C, Berrada F, Poirier J (July 1998). "The neurobiology of apolipoproteins and their receptors in the CNS and Alzheimer's disease". Brain Research. Brain Research Reviews. 27 (2): 119–42. PMID 9622609. doi:10.1016/S0165-0173(98)00008-3.
- Roses AD, Einstein G, Gilbert J, Goedert M, Han SH, Huang D, Hulette C, Masliah E, Pericak-Vance MA, Saunders AM, Schmechel DE, Strittmatter WJ, Weisgraber KH, Xi PT (January 1996). "Morphological, biochemical, and genetic support for an apolipoprotein E effect on microtubular metabolism". Annals of the New York Academy of Sciences. 777 (1): 146–57. PMID 8624078. doi:10.1111/j.1749-6632.1996.tb34413.x.
- Strittmatter WJ, Roses AD (May 1995). "Apolipoprotein E and Alzheimer disease". Proceedings of the National Academy of Sciences of the United States of America. 92 (11): 4725–7. PMC 41779
 . PMID 7761390. doi:10.1073/pnas.92.11.4725.
. PMID 7761390. doi:10.1073/pnas.92.11.4725. - de Knijff P, van den Maagdenberg AM, Frants RR, Havekes LM (1995). "Genetic heterogeneity of apolipoprotein E and its influence on plasma lipid and lipoprotein levels". Human Mutation. 4 (3): 178–94. PMID 7833947. doi:10.1002/humu.1380040303.
- Moriyama K, Sasaki J, Matsunaga A, Arakawa F, Takada Y, Araki K, Kaneko S, Arakawa K (September 1992). "Apolipoprotein E1 Lys-146----Glu with type III hyperlipoproteinemia". Biochimica et Biophysica Acta. 1128 (1): 58–64. PMID 1356443. doi:10.1016/0005-2760(92)90257-V.
- Mahley RW (April 1988). "Apolipoprotein E: cholesterol transport protein with expanding role in cell biology". Science. 240 (4852): 622–30. PMID 3283935. doi:10.1126/science.3283935.
- Utermann G, Pruin N, Steinmetz A (January 1979). "Polymorphism of apolipoprotein E. III. Effect of a single polymorphic gene locus on plasma lipid levels in man". Clinical Genetics. 15 (1): 63–72. PMID 759055. doi:10.1111/j.1399-0004.1979.tb02028.x.
External links
- Apolipoproteins E at the US National Library of Medicine Medical Subject Headings (MeSH)
- apoe4.info - website for APOE-epsilon-4 carriers
- Human APOE genome location and APOE gene details page in the UCSC Genome Browser.