5-Methylcytosine
 | |
| Names | |
|---|---|
| IUPAC name
4-amino-5-methyl-3H-pyrimidin-2-one | |
| Identifiers | |
| 3D model (JSmol) |
|
| ChEBI | |
| ChemSpider | |
| ECHA InfoCard | 100.008.236 |
| KEGG | |
| MeSH | 5-Methylcytosine |
| PubChem CID |
|
| UNII | |
| |
| |
| Properties | |
| C5H7N3O | |
| Molar mass | 125.13 g·mol−1 |
| Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| | |
| Infobox references | |
5-Methylcytosine is a methylated form of the DNA base cytosine that may be involved in the regulation of gene transcription. When cytosine is methylated, the DNA maintains the same sequence, but the expression of methylated genes can be altered (the study of this is part of the field of epigenetics). 5-Methylcytosine is incorporated in the nucleoside 5-methylcytidine.
In 5-methylcytosine, a methyl group is attached to the 5th atom in the 6-atom ring (counting counterclockwise from the NH nitrogen at the six o'clock position, not the 2 o'clock). This methyl group distinguishes 5-methylcytosine from cytosine.
Discovery
While trying to isolate the bacterial toxin responsible for tuberculosis, W.G. Ruppel isolated a novel nucleic acid named tuberculinic acid in 1898 from Tubercle bacillus.[1] The nucleic acid was found to be unusual, in that it contained in addition to thymine, guanine and cytosine, a methylated nucleotide. In 1925, Johnson and Coghill successfully detected a minor amount of a methylated cytosine derivative as a product of hydrolysis of tuberculinic acid with sulfuric acid.[2][3] This report was severely criticized because their identification based solely on the optical properties of the crystalline picrate, and other scientists failed to reproduce the same result.[4] But its existence was ultimately proven in 1948, when Hotchkiss separated the nucleic acids of DNA from calf thymus using paper chromatography, by which he detected a unique methylated cytosine, quite distinct from conventional cytosine and uracil.[5] After seven decades, it turned out that it is also a common feature in different RNA molecules, although the precise role is uncertain.[6]
In vivo
5-Methylcytosine is an epigenetic modification formed by the action of DNA methyltransferases.
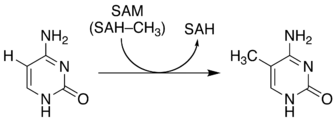
The function of this chemical varies significantly among species:[7]
- In bacteria, 5-methylcytosine can be found at a variety of sites, and is often used as a marker to protect DNA from being cut by native methylation-sensitive restriction enzymes.
- In plants, 5-methylcytosine occurs at CpG, CpHpG and CpHpH sequences (where H = A, C or T).
- In fungi and animals, 5-methylcytosine predominantly occurs at CpG dinucleotides. Most eukaryotes methylate only a small percentage of these sites, but 70-80% of CpG cytosines are methylated in vertebrates.
While spontaneous deamination of cytosine forms uracil, which is recognized and removed by DNA repair enzymes, deamination of 5-methylcytosine forms thymine. This conversion of a DNA base from cytosine (C) to thymine (T) can result in a transition mutation.[8] In addition, active enzymatic deamination of cytosine or 5-methylcytosine by the APOBEC family of cytosine deaminases could have beneficial implications on various cellular processes as well as on organismal evolution.[9] The implications of deamination on 5-hydroxymethylcytosine, on the other hand, remains less understood.
In vitro
The NH2 group can be removed (deamination) from 5-methylcytosine to form thymine with use of reagents such as nitrous acid; cytosine deaminates to uracil under similar conditions.

5-Methylcytosine is resistant to deamination by bisulfite treatment, which deaminates cytosine residues. This property is often exploited to analyze DNA cytosine methylation patterns with bisulfite sequencing.[10]
References
- ↑ Matthews AP (2012). Physiological Chemistry. Williams & Wilkins Company/. p. 167. ISBN 1130145379.
- ↑ Johnson TB, Coghill RD (1925). "The discovery of 5-methyl-cytosine in tuberculinic acid, the nucleic acid of the Tubercle bacillus". J Am Chem Soc. 47 (11): 2838–2844. doi:10.1021/ja01688a030.
- ↑ Grosjean H (2009). Nucleic Acids Are Not Boring Long Polymers of Only Four Types of Nucleotides: A Guided Tour. Landes Bioscience.
- ↑ Vischer E, Zamenhof S, Chargaff E (1949). "Microbial nucleic acids: the desoxypentose nucleic acids of avian tubercle bacilli and yeast". J Biol Chem. 177 (1): 429–438. PMID 18107446.
- ↑ Hotchkiss RD (1948). "The quantitative separation of purines, pyrimidines and nucleosides by paper chromatography". J Biol Chem. 175 (1): 315–332. PMID 18873306.
- ↑ Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T (2012). "Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA". Nucleic Acids Res. 40 (11): 5023–5033. PMC 3367185
 . PMID 22344696. doi:10.1093/nar/gks144.
. PMID 22344696. doi:10.1093/nar/gks144. - ↑ Colot V, Rossignol JL (1999). "Eukaryotic DNA methylation as an evolutionary device". BioEssays. 21 (5): 402–411. PMID 10376011. doi:10.1002/(SICI)1521-1878(199905)21:5<402::AID-BIES7>3.0.CO;2-B.
- ↑ Sassa A, Kanemaru Y, Kamoshita N, Honma M, Yasui M (2016). "Mutagenic consequences of cytosine alterations site-specifically embedded in the human genome". Genes and Environ. 38 (1): 17. PMC 5007816
 . PMID 27588157. doi:10.1186/s41021-016-0045-9.
. PMID 27588157. doi:10.1186/s41021-016-0045-9. - ↑ Chahwan R, Wontakal SN, Roa S (2010). "Crosstalk between genetic and epigenetic information through cytosine deamination". Trends in Genetics. 26 (10): 443–448. PMID 20800313. doi:10.1016/j.tig.2010.07.005.
- ↑ Clark SJ, Harrison J, Paul CL, Frommer M (1994). "High sensitivity mapping of methylated cytosines". Nucleic Acids Res. 22 (15): 2990–2997. PMC 310266
 . PMID 8065911. doi:10.1093/nar/22.15.2990.
. PMID 8065911. doi:10.1093/nar/22.15.2990.
Literature
- Griffiths, Anthony J. F. (1999). An Introduction to genetic analysis. San Francisco: W.H. Freeman. Chapter 15: Gene Mutation. ISBN 0-7167-3520-2. (available online at the United States National Center for Biotechnology Information)