Viral metagenomics
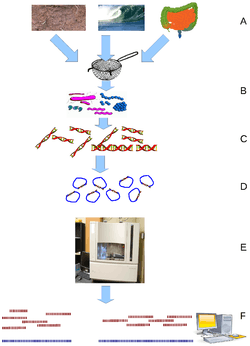
Environmental Shotgun Sequencing (ESS). (A) Sampling from habitat; (B) filtering particles, typically by size; (C) Lysis and DNA extraction; (D) cloning and library construction; (E) sequencing the clones; (F) sequence assembly into contigs and scaffolds.
Viral metagenomics is the study of viral genetic material sourced directly from the environment rather than from a host or natural reservoir. The goal is to ascertain viral diversity in the environment that is often missed in studies targeting specific potential reservoirs. It reveals important information on virus evolution and the genetic diversity of the viral community without the need for isolating viral species and cultivating them in the laboratory. This approach has created improvements in molecular epidemiology and accelerated the discovery of novel viruses.[1][2]
See also
References
- ↑ David M. Kristensen1, Arcady R. Mushegian1, 2, Valerian V. Dolja3 and Eugene V. Koonin4, New dimensions of the virus world discovered through metagenomics. Trends in Microbiology, Volume 18, Issue 1, 11-19, 25 November 2009.
- ↑ Bernardo P, Albina E, Eloit M, Roumagnac P.. Pathology and viral metagenomics, a recent history.[Article in French].Med Sci (Paris). 2013 May;29(5):501-8.
External links
- CAMERA Cyberinfrastructure for Metagenomics, data repository and tools for metagenomics research.
- GOLD Genomes OnLine Database (GOLD).
- IMG/M The Integrated Microbial Genomes system, for metagenome analysis by the DOE-JGI.
- MEGAN MEtaGenome ANalyzer. A stand-alone metagenome analysis tool.
- MetaGeneMark MetaGeneMark for MetaGenome Gene Finding
- Metagenomics and Our Microbial Planet A website on metagenomics and the vital role of microbes on Earth from the National Academies.
- Metagenomics at the European Bioinformatics Institute Analysis and archiving of metagenomic data.
- Metagenomics: Sequences from the Environment free ebook from NCBI Bookshelf.
- MG-RAST Metagenomics Rapid Annotation using Subsystem Technology server
- The New Science of Metagenomics: Revealing the Secrets of Our Microbial Planet A report released by the National Research Council in March 2007. Also, see the Report In Brief.
| ||||||||||||||||||||||
This article is issued from Wikipedia - version of the Wednesday, September 30, 2015. The text is available under the Creative Commons Attribution/Share Alike but additional terms may apply for the media files.