Transposons as a genetic tool
Transposons are semi-parasitic DNA sequences which can replicate and spread through the host's genome. They can be harnessed as a genetic tool for analysis of gene and protein function. The use of transposons is well-developed in Drosophila (in which P elements are most commonly used) and in Thale cress (Arabidopsis thaliana) and bacteria such as Escherichia coli (E. coli ).[1][2]
Currently transposons can be used in genetic research and recombinant genetic engineering for insertional mutagenesis. Insertional mutagenesis is when transposons function as vectors to help remove and integrate genetic sequences. Given their relatively simple design and inherent ability to move DNA sequences, transposons are highly compatible at transducing genetic material, making them ideal genetic tools.
Signature-Tagging Mutagenesis
Signature-tagging mutagenesis (also known as STM) is a technique focused on using transposable element insertion to determine the phenotype of a locus in an organism's genome. While genetic sequencing techniques can determine the genotype of a genome, they cannot determine the function or phenotypic expression of gene sequences.[3][4] STM can bypass this issue by mutating a locus, causing it form a new phenotype; by comparing the observed phenotypic expressions of the mutated and unaltered locus, one can deduce the phenotypic expression of the locus.
In STM, specially tagged transposons are inserted into an organism, such as a bacterium, and randomly integrated into the host genome. In theory, the modified mutant organism should express the altered gene, thus altering the phenotype. If a new phenotype is observed, the genome is sequenced and searched for tagged transposons.[3] If the site of transposon integration is found, then the locus may be responsible for expressing the phenotypes.[5][6]
There have been many studies conducted transposon based STM, most notably with the P elements[4] in Drosophila. P elements are transposons originally described in Drosophila melanogaster genome capable of being artificially synthesized or spread to other Drosophila species through horizontal transfer.[4] In experimental trials, artificially created P elements and transposase genes are inserted into the genomes of Drosophila embryos. Subsequently, embryos that exhibit mutations have their genomes sequenced and compared, thus revealing the loci that have been affected by insertion and the roles of the loci.,[4][6]
Insertional Inactivation
Insertional inactivation focuses on suppressing the expression of a gene by disrupting its sequence with an insertion. When additional nucleotides are inserted near or into a locus, the locus can suffer a frameshift mutation that could prevent it from being properly expressed into polypeptide chain. Transposon-based Insertional inactivation is considered for medical research from suppression of antibiotic resistance in bacteria to the treatment of genetic diseases.[7] In the treatment of genetic diseases, the insertion of a transposon into deleterious gene locus of organism's genome would misalign the locus sequence, truncating any harmful proteins formed and rendering them non-functional. Alternatively insertional inactivation could be used to suppress genes that express antibiotic-resistance in bacteria.,[7][8]
Sleeping Beauty
While transposons have been used successfully in plants and invertebrate subjects through insertional mutagenesis and insertional activation, the usage of transposons in vertebrates has been limited due to a lack of transposons specific to vertebrates. Nearly all transposons compatible to and present within vertebrate genomes are inactive and are often relegated to "junk" DNA.[6] However it is possible to identify dormant transposons and artificially recreate them as active agents.[6] Genetic researchers Zsuzsanna Izsvák and Zoltán Ivics discovered a fish transposon sequence that, despite being dormant for 15 million years, could be resurrected as a vector for introducing foreign genes into the vertebrate genomes, including those of humans. This transposon, called Sleeping Beauty was described in 1997, and could be artificially reactivated into a functioning transposon.[6]
Sleeping Beauty can also be viable in gene therapy procedures by helping introduce beneficial transgenes into host genomes. Belcher et al. tested this notion by using Sleeping Beauty transposons to help insert sequences into mice with sickle cell anemia so they can produce the enzymes need to counteract their anemia.[6] Belcher et al. began their experiment by constructing a genetic sequence consisting of the Hmox-1 transposable element and transposase from Sleeping Beauty. This sequence was then added inserted into a plasmid and introduced into the cells of the mice. The transposase from Sleeping Beauty helped insert the Hmox-1 transposon into the mice genome, allowing the production of enzyme heme oxygenase-1 (HO-1). The mice that receive the insertion showed a fivefold increase in the expression of HO-1, which in turn reduced blood vessel blockage from sickle-cell anemia. The publication of the experiment in 2010 showed that transposons can be useful in gene therapy.[6]
P Elements as a tool (Drosophila)
Naturally occurring P elements contain:
- coding sequence for the enzyme transposase;
- recognition sequences for transposase action.
Transposase is an enzyme which regulates and catalyzes the excision of a P element from the host DNA, cutting at two recognition sites, and then reinserts the P element randomly. It is the random-insertion process, that can interfere with existing genes, or carry an additional gene, that can be used as a process for genetic research.
To use this process as a useful and controllable genetic tool, the two parts of the P element must be separated to prevent uncontrolled transposition. The normal genetic tools are therefore:
- DNA coding for transposase (or occasisonally simply transposase) with no transposase recognition sequences so it cannot insert; and
- a "P Plasmid".
P Plasmids always contain:
- a Drosophila reporter gene, often a red-eye marker (the product of the white gene);
- transposase recognition sequences;
and may contain:
- a gene of interest;
- an E. coli reporter gene (often some kind of antibiotic resistance); and
- origin of replication and other associated plasmid 'housekeeping' sequences.
Methods of usage (Drosophila)
(Forward genetics methods) There are two main ways to utilise these tools: Fly Transformation, and Insertional Mutagenesis, each described below.
Fly Transformation
(hoping for insertion in non-coding regions)
- Microinject the posterior end of an early-stage (pre-cellularization) embryo with coding for transposase and a plasmid with the reporter gene, gene of interest and transposase recognition sequences.
- Random transposition occurs, inserting the gene of interest and reporter gene.
- Grow flies and cross to remove genetic variation between the cells of the organism. (Only some of the cells of the organism will have been transformed. By breeding only the genotype of the gametes is passed on, removing this variation).
- Look for flies expressing the reporter gene. These carry the inserted gene of interest, so can be investigated to determine the phenotype due to the gene of interest.
It is important to note that the inserted gene may have damaged the function of one of the host's genes. Several lines of flies are required so comparison can take place and ensure that no additional genes have been knocked out.
Insertional Mutagenesis
(hoping for insertion in coding region)
- Microinject the embryo with coding for transposase and a plasmid with the reporter gene and transposase recognition sequences (and often the E. coli reporter gene and origin of replication, etc.).
- Random transposition occurs, inserting the reporter gene randomly. The insertion tends to occur near actively transcribed genes, as this is where the chromatin structure is loosest, so the DNA most accessible.
- Grow flies and cross to remove genetic variation between the cells of the organism (see above).
- Look for flies expressing the reporter gene. These have experienced a successful transposition, so can be investigated to determine the phenotype due to mutation of existing genes.
Possible mutations:
- Insertion in a translated region => hybrid protein/truncated protein. Usually causes loss of protein function, although more complex effects are seen.
- Insertion in an intron => altered splicing pattern/splicing failure. Usually results in protein truncation or the production of inactive mis-spliced products, although more complex effects are common.
- Insertion in 5' (the sequence that will become the mRNA 5' UTR) untranslated region => truncation of transcript. Usually results in failure of the mRNA to contain a 5' cap, leading to less efficient translation.
- Insertion in promoter => reduction/complete loss of expression. Always results in greatly reduced protein production levels. The most useful type of insertion for analysis due to the simplicity of the situation.
- Insertion between promoter and upstream enhancers => loss of enhancer function/hijack of enhancer function for reporter gene.† Generally reduces the level of protein specificity to cell type, although complex effects are often seen.
† Enhancer trapping
The hijack of an enhancer from another gene allows the analysis of the function of that enhancer. This, especially if the reporter gene is for a fluorescent protein, can be used to help map expression of the mutated gene through the organism, and is a very powerful tool.
Other usage of P Elements (Drosophila)
(Reverse genetics method)
Secondary mobilisation
If there is an old P element near the gene of interest (with a broken transposase) you can remobilise by microinjection of the embryo with coding for transposase or transposase itself. The P element will often transpose within a few kilobases of the original location, hopefully affecting your gene of interest as for 'Insertional Mutagenisis'.
Analysis of Mutagenesis Products (Drosophila)
Once the function of the mutated protein has been determined it is possible to sequence/purify/clone the regions flanking the insertion by the following methods:
Inverse PCR
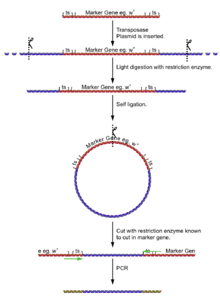
Main article: Inverse PCR
- Isolate the fly genome.
- Undergo a light digest (using an enzyme [enzyme 1] known NOT to cut in the reporter gene), giving fragments of a few kilobases, a few with the insertion and its flanking DNA.
- Self ligate the digest (low DNA concentration to ensure self ligation) giving a selection of circular DNA fragments, a few with the insertion and its flanking DNA.
- Cut the plasmids at some point in the reporter gene (with an enzyme [enzyme 2] known to cut very rarely in genomic DNA, but is known to in the reporter gene).
- Using primers for the reporter gene sections, the DNA can be amplified for sequencing.
The process of cutting, self ligation and re cutting allows the amplification of the flanking regions of DNA without knowing the sequence. The point at which the ligation occurred can be seen by identifying the cut site of [enzyme 1].
Plasmid Rescue (E. coli Transformation)

- Isolate the fly genome.
- Undergo a light digest (using an enzyme [enzyme 1] known to cut in the boundary between the reporter gene and the E. coli reporter gene and plasmid sequences), giving fragments of a few kilobases, a few with the E. coli reporter, the plasmid sequences and its flanking DNA.
- Self ligate the digest (low DNA concentration to ensure self ligation) giving a selection of circular DNA fragments, a few with the E. coli reporter, the plasmid sequences and its flanking DNA.
- Insert the plasmids into E. coli cells (e.g. by electroporation).
- Screen plasmids for the E. coli reporter gene. Only successful inserts of plasmids with the plasmid 'housekeeping' sequences will express this gene.
7. The gene can be cloned for further analysis.
Transposable Element Application Other organisms
The genomes of other organisms can be analysed in a similar way, although with different transposable elements. The recent discovery of the 'mariner transposon' (from the reconstruction of the original sequence from many 'dead' versions in the human genome) has allowed many new experiments, mariner has well conserved homologues across a wide range of species and is a very versatile tool.
References
This article incorporates material from the Citizendium article "Transposons as a genetic tool", which is licensed under the Creative Commons Attribution-ShareAlike 3.0 Unported License but not under the GFDL.
- ↑ Beckwith, J.; Silhavy, T.J. (1992). The Power of Bacterial Genetics: A Literature Based Course. NY: Cold Spring Harbor Laboratory Press. pp. 555–614. ISBN 0-87969-411-4.
- ↑ Kleckner N, Roth J, Botstein D (October 1977). "Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics". J. Mol. Biol. 116 (1): 125–59. doi:10.1016/0022-2836(77)90123-1. PMID 338917.
- 1 2 Berg, Claire; Berg, Douglass E. "Transposable Elements Tools for Microbial Genetics". EcoSal.
- 1 2 3 4 Engels, William R. "P Elements in Drosophila". Genetics Department, University of Wisconsin.
- ↑ Ivics Z, Li MA, Mátés L, et al. (June 2009). "Transposon-mediated genome manipulation in vertebrates". Nat. Methods 6 (6): 415–22. doi:10.1038/nmeth.1332. PMC 2867038. PMID 19478801.
- 1 2 3 4 5 6 7 Luft FC (July 2010). "Sleeping Beauty jumps to new heights". J. Mol. Med. 88 (7): 641–3. doi:10.1007/s00109-010-0626-1. PMID 20467721.
- 1 2 Berger-Bächi B (April 1983). "Insertional inactivation of staphylococcal methicillin resistance by Tn551". J. Bacteriol. 154 (1): 479–87. PMC 217482. PMID 6300037.
- ↑ Zoltaná Ivics; Meng Amy Li; Lajos Mates; Jef D. Boeke; Allan Bradley;. "Transposon-mediated Genome Manipulations in Vertebrates". National Center for Biotechnology Information.
Further reading
- Snyder, L.; Champness, W. (2003). "Ch. 9: Transposition and site-specific recombination". Molecular Genetics of Bacteria (2nd ed.). Washington DC: ASM Press. pp. 303–340. ISBN 1-55581-204-X.
- "Bibliography: Transposons as a genetic tool". Citizendium.org.
- See Martienssen, R.A.; Springer, P.S. "Enhancer and Gene Trap Transposon Mutagenesis in Arabidopsis". Cold Spring Harbor Laboratory.