SCNN1B
The SCNN1B gene encodes for the β subunit of the epithelial sodium channel ENaC in vertebrates. ENaC is assembled as a heterotrimer composed of three homologous subunits α, β, and γ or δ, β, and γ. The other ENAC subunits are encoded by SCNN1A, SCNN1G, and SCNN1D.[1]
ENaC is expressed in epithelial cells[1] and is different from the voltage-gated sodium channel that is involved in the generation of action potentials in neurons. The abbreviation for the genes encoding for voltage-gated sodium channel starts with three letters: SCN. In contrast to these sodium channels, ENaC is constitutively active and is not voltage-dependent. The second N in the abbreviation (SCNN1A) represents that these are NON-voltage-gated channels.
In most vertebrates, sodium ions are the major determinant of the osmolarity of the extracellular fluid.[2] ENaC allows transfer of sodium ions across the epithelial cell membrane in so-called "tight-epithelia" that have low permeability. The flow of sodium ions across epithelia affects osmolarity of the extracellular fluid. Thus, ENaC plays a central role in the regulation of body fluid and electrolyte homeostasis and consequently affects blood pressure.[3]
As ENaC is strongly inhibited by amiloride, it is also referred to as an "amiloride-sensitive sodium channel".
History
The first cDNA encoding the beta subunit of ENaC was cloned and sequenced by Canessa et al. from rat mRNA.[4] A year later, two independent groups reported the cDNA sequences of the beta- and gamma-subunits of the human ENaC.[5][6] The exon-intron organization of the human beta ENaC gene SCNN1B was determined by Saxena et al. by sequencing genomic DNA from three subjects from three different ethnic groups.[7] This study also established that the exon-intron architecture of the three subunits of ENaC have remained highly conserved despite the divergence of their sequences.[7]
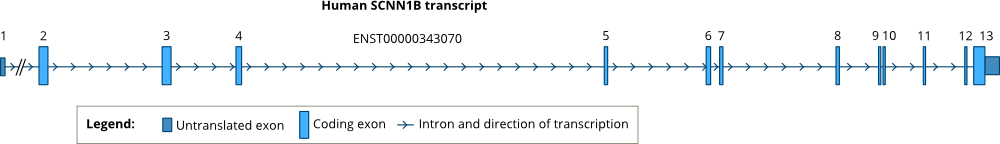
Gene structure
While the human gene SCNN1A is located in chromosome 12p,[8] the human genes encoding SCNN1B and SCNN1G are located in juxtaposition in the short arm of chromosome 16 (16p12-p13).[6] Sequencing of the human genomic DNA indicated that the SCNN1B gene has 13 exons separated by 12 introns.[7] The positions of introns are conserved in all three human ENaC genes, SCNN1A, SCNN1B and SCNN1G.[7] The positions of the introns are also highly conserved across vertebrates. See: Ensembl GeneTree.
Analysis of transcripts of the SCNN1B gene in human kidney and lung showed several alternative transcription and translation initiation sites.[9] However, only one of these transcripts (ENST00000343070) is highly expressed and other transcripts appear at low amounts.[9]
Tissue-specific expression
The three ENaC subunits encoded by SCNN1A, SCNN1B, and SCNN1G are commonly expressed in tight epithelia that have low water permeability.[1] The major organs where ENaC is expressed include parts of the kidney tubular epithelia,[3][10] the respiratory airway,[11] the female reproductive tract,[11] colon, salivary and sweat glands.[10]
ENaC is also expressed in the tongue, where it has been shown to be essential for the perception of salt taste.[10]
The expression of ENaC subunit genes is regulated mainly by the mineralocorticoid hormone aldosterone that is activated by the renin-angiotensin system.[12] [13]
Protein structure
The primary structures of all four ENaC subunits show strong similarity. Thus, these four proteins represent a family of proteins that share a common ancestor. In global alignment (meaning alignments of sequences along their entire length and not just a partial segment), the human β subunit shares 34% identity with the γ subunit and 26 and 23% identity with the α and δ subunits.[1]
All four ENaC subunit sequences have two hydrophobic stretches that form two transmembrane segments named as TM1 and TM2.[14] In the membrane-bound form, the TM segments are embedded in the membrane bilayer, the amino- and carboxy-terminal regions are located inside the cell, and the segment between the two TMs remains outside of the cell as the extracellular region of ENaC. This extracellular region includes about 70% of the residues of each subunit. Thus, in the membrane-bound form, the bulk of each subunit is located outside of the cell.
The structure of ENaC has not been yet determined. Yet, the structure of a homologous protein ASIC1 has been resolved.[15][16] The chicken ASIC1 structure revealed that ASIC1 is assembled as a homotrimer of three identical subunits. The authors of the original study suggested that the ASIC1 trimer resembles a hand holding a ball.[15] Hence distinct domains of ASIC1 have been referred to as palm, knuckle, finger, thumb, and β-ball.[15]
Alignment of ENaC subunit sequences with ASIC1 sequence reveals that TM1 and TM2 segments and palm domain are conserved, and the knuckle, finger and thumb domains have insertions in ENaC. Site-directed mutagenesis studies on ENaC subunits provide evidence that many basic features of the ASIC1 structural model apply to ENaC as well.
In the carboxy terminus of three ENaC subunits, (α, β and γ) there is a special conserved consensus sequence PPPXYXXL that is called the PY motif. This sequence is recognized by the so-called WW domains in a special E3 ubiquitin-protein ligase named Nedd4-2.[17] Nedd4-2 ligates ubiquitin to the C-terminus of the ENaC subunit which marks the protein for degradation.[17]
Associated diseases
At present, three major hereditary disorders are known to be associated with mutations in the SCNN1B gene. These are: 1. Multisystem pseudohypoaldosteronism, 2. Liddle syndrome, and 3. Cystic fibrosis-like disease.[1]
Multi-system form of type I pseudohypoaldosteronism (PHA1B)
The disease most commonly associated with mutations in SCNN1B is the multi-system form of type I pseudohypoaldosteronism (PHA1B) that was first characterized by A. Hanukoglu as an autosomal recessive disease.[18] This is a syndrome of unresponsiveness to aldosterone in patients that have high serum levels of aldosterone but suffer from symptoms of aldosterone deficiency with a high risk of mortality due to severe salt loss. Initially, this disease was thought to be a result of a mutation in the mineralocorticoid receptor (NR3C2) that binds aldosterone. But homozygosity mapping in 11 affected families revealed that the disease is associated with two loci on chromosome 12p13.1-pter and chromosome 16p12.2-13 that include the genes for SCNN1A and SCNN1B and SCNN1G respectively.[19] Sequencing of the ENaC genes identified mutation in affected patients, and functional expression of the mutated cDNAs further confirmed that identified mutations lead to the loss of activity of ENaC.[20]
In the majority of the patients with multi-system PHA1B a homozygous mutation or two compound heterozygous mutations have been detected.[21][22][23]
Liddle syndrome
Liddle syndrome is generally caused by mutations in the PY motif or truncation of the C-terminus including loss of the PY motif in the β or γ ENaC subunits.[24][25][26][27][28][29] Even though there is a PY motif also in the α subunit, so far Liddle disease has not observed in association with a mutation in the α subunit. Liddle syndrome is inherited as an autosomal dominant disease with a phenotype that includes early onset hypertension, metabolic alkalosis and low levels of plasma renin activity and mineralocorticoid hormone aldosterone. In the absence of a recongnizable PY motif, ubiquitin-protein ligase Nedd4-2 cannot bind to the ENaC subunit and hence cannot attach a ubiquitin to it. Consequently, proteolysis of ENaC by proteasome is inhibited and ENaC accumulates in the membrane leading to enhanced activity of ENaC that causes hypertension.[30][31][32][33]
Interactions
SCNN1B has been shown to interact with WWP2[34][35] and NEDD4.[34][35][36]
References
- 1 2 3 4 5 Hanukoglu I, Hanukoglu A (Jan 2016). "Epithelial sodium channel (ENaC) family: Phylogeny, structure-function, tissue distribution, and associated inherited diseases.". Gene 579 (2): 95–132. doi:10.1016/j.gene.2015.12.061. PMID 26772908.
- ↑ Bourque CW (Jul 2008). "Central mechanisms of osmosensation and systemic osmoregulation". Nature Reviews. Neuroscience 9 (7): 519–31. doi:10.1038/nrn2400. PMID 18509340.
- 1 2 Rossier BC, Baker ME, Studer RA (Jan 2015). "Epithelial sodium transport and its control by aldosterone: the story of our internal environment revisited". Physiological Reviews 95 (1): 297–340. doi:10.1152/physrev.00011.2014. PMID 25540145.
- ↑ Canessa CM, Schild L, Buell G, Thorens B, Gautschi I, Horisberger JD, Rossier BC (Feb 1994). "Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits.". Nature 367 (6462): 463–7. doi:10.1038/367463a0. PMID 8107805.
- ↑ McDonald FJ, Price MP, Snyder PM, Welsh MJ (May 1995). "Cloning and expression of the beta- and gamma-subunits of the human epithelial sodium channel". American Journal of Physiology 268 (5 Pt 1): C1157–63. PMID 7762608.
- 1 2 Voilley N, Bassilana F, Mignon C, Merscher S, Mattéi MG, Carle GF, Lazdunski M, Barbry P (Aug 1995). "Cloning, chromosomal localization, and physical linkage of the beta and gamma subunits (SCNN1B and SCNN1G) of the human epithelial amiloride-sensitive sodium channel". Genomics 28 (3): 560–5. doi:10.1006/geno.1995.1188. PMID 7490094.
- 1 2 3 4 Saxena A, Hanukoglu I, Strautnieks SS, Thompson RJ, Gardiner RM, Hanukoglu A (Nov 1998). "Gene structure of the human amiloride-sensitive epithelial sodium channel beta subunit". Biochemical and Biophysical Research Communications 252 (1): 208–13. doi:10.1006/bbrc.1998.9625. PMID 9813171.
- ↑ Ludwig M, Bolkenius U, Wickert L, Marynen P, Bidlingmaier F (May 1998). "Structural organisation of the gene encoding the alpha-subunit of the human amiloride-sensitive epithelial sodium channel". Human Genetics 102 (5): 576–81. PMID 9654208.
- 1 2 Thomas CP, Loftus RW, Liu KZ, Itani OA (May 2002). "Genomic organization of the 5' end of human beta-ENaC and preliminary characterization of its promoter". American Journal of Physiology. Renal Physiology 282 (5): F898–909. PMID 11934701.
- 1 2 3 Duc C, Farman N, Canessa CM, Bonvalet JP, Rossier BC (Dec 1994). "Cell-specific expression of epithelial sodium channel alpha, beta, and gamma subunits in aldosterone-responsive epithelia from the rat: localization by in situ hybridization and immunocytochemistry". The Journal of Cell Biology 127 (6 Pt 2): 1907–21. PMID 7806569.
- 1 2 Enuka Y, Hanukoglu I, Edelheit O, Vaknine H, Hanukoglu A (Mar 2012). "Epithelial sodium channels (ENaC) are uniformly distributed on motile cilia in the oviduct and the respiratory airways". Histochemistry and Cell Biology 137 (3): 339–53. doi:10.1007/s00418-011-0904-1. PMID 22207244.
- ↑ Palmer LG, Patel A, Frindt G (Feb 2012). "Regulation and dysregulation of epithelial Na+ channels". Clinical and Experimental Nephrology 16 (1): 35–43. doi:10.1007/s10157-011-0496-z. PMID 22038262.
- ↑ Thomas W, Harvey BJ (2011). "Mechanisms underlying rapid aldosterone effects in the kidney". Annual Review of Physiology 73: 335–57. doi:10.1146/annurev-physiol-012110-142222. PMID 20809792.
- ↑ Canessa CM, Merillat AM, Rossier BC (Dec 1994). "Membrane topology of the epithelial sodium channel in intact cells". The American Journal of Physiology 267 (6 Pt 1): C1682–90. PMID 7810611.
- 1 2 3 Jasti J, Furukawa H, Gonzales EB, Gouaux E (Sep 2007). "Structure of acid-sensing ion channel 1 at 1.9 A resolution and low pH". Nature 449 (7160): 316–23. doi:10.1038/nature06163. PMID 17882215.
- ↑ Baconguis I, Bohlen CJ, Goehring A, Julius D, Gouaux E (Feb 2014). "X-ray structure of acid-sensing ion channel 1-snake toxin complex reveals open state of a Na(+)-selective channel". Cell 156 (4): 717–29. doi:10.1016/j.cell.2014.01.011. PMID 24507937.
- 1 2 Rotin D, Staub O (Jan 2011). "Role of the ubiquitin system in regulating ion transport". Pflugers Archiv : European journal of physiology 461 (1): 1–21. doi:10.1007/s00424-010-0893-2. PMID 20972579.
- ↑ Hanukoglu A (Nov 1991). "Type I pseudohypoaldosteronism includes two clinically and genetically distinct entities with either renal or multiple target organ defects". The Journal of Clinical Endocrinology and Metabolism 73 (5): 936–44. doi:10.1210/jcem-73-5-936. PMID 1939532.
- ↑ Strautnieks SS, Thompson RJ, Hanukoglu A, Dillon MJ, Hanukoglu I, Kuhnle U, Seckl J, Gardiner RM, Chung E (Feb 1996). "Localisation of pseudohypoaldosteronism genes to chromosome 16p12.2-13.11 and 12p13.1-pter by homozygosity mapping". Human Molecular Genetics 5 (2): 293–9. PMID 8824886.
- ↑ Chang SS, Grunder S, Hanukoglu A, Rösler A, Mathew PM, Hanukoglu I, Schild L, Lu Y, Shimkets RA, Nelson-Williams C, Rossier BC, Lifton RP (Mar 1996). "Mutations in subunits of the epithelial sodium channel cause salt wasting with hyperkalaemic acidosis, pseudohypoaldosteronism type 1". Nature Genetics 12 (3): 248–53. doi:10.1038/ng0396-248. PMID 8589714.
- ↑ Strautnieks SS, Thompson RJ, Gardiner RM, Chung E (Jun 1996). "A novel splice-site mutation in the gamma subunit of the epithelial sodium channel gene in three pseudohypoaldosteronism type 1 families". Nature Genetics 13 (2): 248–50. doi:10.1038/ng0696-248. PMID 8640238.
- ↑ Edelheit O, Hanukoglu I, Gizewska M, Kandemir N, Tenenbaum-Rakover Y, Yurdakök M, Zajaczek S, Hanukoglu A (May 2005). "Novel mutations in epithelial sodium channel (ENaC) subunit genes and phenotypic expression of multisystem pseudohypoaldosteronism". Clinical Endocrinology 62 (5): 547–53. doi:10.1111/j.1365-2265.2005.02255.x. PMID 15853823.
- ↑ Zennaro MC, Hubert EL, Fernandes-Rosa FL (Mar 2012). "Aldosterone resistance: structural and functional considerations and new perspectives". Molecular and Cellular Endocrinology 350 (2): 206–15. doi:10.1016/j.mce.2011.04.023. PMID 21664233.
- ↑ Hansson JH, Nelson-Williams C, Suzuki H, Schild L, Shimkets R, Lu Y, Canessa C, Iwasaki T, Rossier B, Lifton RP (1995). "Hypertension caused by a truncated epithelial sodium channel gamma subunit: genetic heterogeneity of Liddle syndrome". Nat. Genet. 11 (1): 76–82. doi:10.1038/ng0995-76. PMID 7550319.
- ↑ Shimkets RA, Warnock DG, Bositis CM, Nelson-Williams C, Hansson JH, Schambelan M, Gill JR, Ulick S, Milora RV, Findling JW (1994). "Liddle's syndrome: heritable human hypertension caused by mutations in the beta subunit of the epithelial sodium channel". Cell 79 (3): 407–14. doi:10.1016/0092-8674(94)90250-X. PMID 7954808.
- ↑ Hansson JH, Schild L, Lu Y, Wilson TA, Gautschi I, Shimkets R, Nelson-Williams C, Rossier BC, Lifton RP (1996). "A de novo missense mutation of the beta subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity". Proc. Natl. Acad. Sci. U.S.A. 92 (25): 11495–9. doi:10.1073/pnas.92.25.11495. PMC 40428. PMID 8524790.
- ↑ Inoue J, Iwaoka T, Tokunaga H, Takamune K, Naomi S, Araki M, Takahama K, Yamaguchi K, Tomita K (1998). "A family with Liddle's syndrome caused by a new missense mutation in the beta subunit of the epithelial sodium channel". J. Clin. Endocrinol. Metab. 83 (6): 2210–3. doi:10.1210/jc.83.6.2210. PMID 9626162.
- ↑ Persu A, Barbry P, Bassilana F, Houot AM, Mengual R, Lazdunski M, Corvol P, Jeunemaitre X (1998). "Genetic analysis of the beta subunit of the epithelial Na+ channel in essential hypertension". Hypertension 32 (1): 129–37. doi:10.1161/01.hyp.32.1.129. PMID 9674649.
- ↑ Uehara Y, Sasaguri M, Kinoshita A, Tsuji E, Kiyose H, Taniguchi H, Noda K, Ideishi M, Inoue J, Tomita K, Arakawa K (1998). "Genetic analysis of the epithelial sodium channel in Liddle's syndrome". J. Hypertens. 16 (8): 1131–5. doi:10.1097/00004872-199816080-00008. PMID 9794716.
- ↑ Snyder PM, Price MP, McDonald FJ, Adams CM, Volk KA, Zeiher BG, Stokes JB, Welsh MJ (1996). "Mechanism by which Liddle's syndrome mutations increase activity of a human epithelial Na+ channel". Cell 83 (6): 969–78. doi:10.1016/0092-8674(95)90212-0. PMID 8521520.
- ↑ Tamura H, Schild L, Enomoto N, Matsui N, Marumo F, Rossier BC (1996). "Liddle disease caused by a missense mutation of beta subunit of the epithelial sodium channel gene". J. Clin. Invest. 97 (7): 1780–4. doi:10.1172/JCI118606. PMC 507244. PMID 8601645.
- ↑ Firsov D, Schild L, Gautschi I, Mérillat AM, Schneeberger E, Rossier BC (1997). "Cell surface expression of the epithelial Na channel and a mutant causing Liddle syndrome: A quantitative approach". Proc. Natl. Acad. Sci. U.S.A. 93 (26): 15370–5. doi:10.1073/pnas.93.26.15370. PMC 26411. PMID 8986818.
- ↑ Pirozzi G, McConnell SJ, Uveges AJ, Carter JM, Sparks AB, Kay BK, Fowlkes DM (1997). "Identification of novel human WW domain-containing proteins by cloning of ligand targets". J. Biol. Chem. 272 (23): 14611–6. doi:10.1074/jbc.272.23.14611. PMID 9169421.
- 1 2 McDonald FJ, Western AH, McNeil JD, Thomas BC, Olson DR, Snyder PM (September 2002). "Ubiquitin-protein ligase WWP2 binds to and downregulates the epithelial Na(+) channel". Am. J. Physiol. Renal Physiol. 283 (3): F431–6. doi:10.1152/ajprenal.00080.2002. PMID 12167593.
- 1 2 Harvey KF, Dinudom A, Cook DI, Kumar S (March 2001). "The Nedd4-like protein KIAA0439 is a potential regulator of the epithelial sodium channel". J. Biol. Chem. 276 (11): 8597–601. doi:10.1074/jbc.C000906200. PMID 11244092.
- ↑ Farr TJ, Coddington-Lawson SJ, Snyder PM, McDonald FJ (February 2000). "Human Nedd4 interacts with the human epithelial Na+ channel: WW3 but not WW1 binds to Na+-channel subunits". Biochem. J. 345 Pt 3 (3): 503–9. doi:10.1042/0264-6021:3450503. PMC 1220784. PMID 10642508.
Further reading
- Alvarez de la Rosa D, Canessa CM, Fyfe GK, Zhang P (2000). "Structure and regulation of amiloride-sensitive sodium channels". Annu. Rev. Physiol. 62: 573–94. doi:10.1146/annurev.physiol.62.1.573. PMID 10845103.
- Rossier BC, Pradervand S, Schild L, Hummler E (2002). "Epithelial sodium channel and the control of sodium balance: interaction between genetic and environmental factors". Annu. Rev. Physiol. 64: 877–97. doi:10.1146/annurev.physiol.64.082101.143243. PMID 11826291.
External links
- SCNN1B protein, human at the US National Library of Medicine Medical Subject Headings (MeSH)
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
