Polymerase cycling assembly
Polymerase cycling assembly (or PCA, also known as Assembly PCR) is a method for the assembly of large DNA oligonucleotides from shorter fragments. The process uses the same technology as PCR, but takes advantage of DNA hybridization and annealing as well as DNA polymerase to amplify a complete sequence of DNA in a precise order based on the single stranded oligonucleotides used in the process. It thus allows for the production of synthetic genes and even entire synthetic genomes.
PCA principles
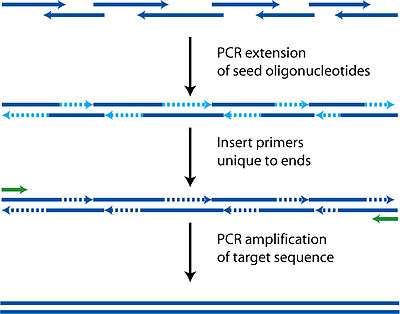
Much like how primers are designed such that there is a forward primer and a reverse primer capable of allowing DNA polymerase to fill the entire template sequence, PCA uses the same technology but with multiple oligonucleotides. While in PCR the customary size of oligonuleotides used is 18 base pairs, in PCA lengths of up to 50 are used to ensure uniqueness and correct hybridization.
Each oligonucleotide is designed to be either part of the top or bottom strand of the target sequence. As well as the basic requirement of having to be able to tile the entire target sequence, these oligonucleotides must also have the usual properties of similar melting temperatures, hairpin free, and not too GC rich to avoid the same complications as PCR.
During the polymerase cycles, the oligonucleotides anneal to complementary fragments and then are filled in by polymerase. Each cycle thus increases the length of various fragments randomly depending on which oligonucleotides find each other. It is critical that there is complementarity between all the fragments in some way or a final complete sequence will not be produced as polymerase requires a template to follow.
After this initial construction phase, additional primers encompassing both ends are added to perform a regular PCR reaction, amplifying the target sequence away from all the shorter incomplete fragments. A gel purification can then be used to identify and isolate the complete sequence.
Typical Reaction
A typical reaction consists of oligonucleotides ~50 base pairs long each overlapping by about 20 base pairs. The reaction with all the oligonucleotides is then carried out for ~30 cycles followed by an additional 23 cycles with the end primers.
Gibson Assembly
A modification of this method, Gibson assembly, described by Gibson et al. allows for single-step isothermal assembly of DNA with up to several hundreds kb. By using T5 exonuclease to 'chew back' complementary ends, an overlap of about 40bp can be created. The reaction takes place at 50°C, a temperature where the T5 exonuclease is unstable. After a short timestep it is degraded, the overlapps can anneal and be ligated.[1] Cambridge University IGEM team made a video describing the process.[2] Ligation independent cloning (LIC) is a new variant of the method for compiling several DNA pieces together and needing only exonuclease enzyme for the reaction. However, the method requires even number of DNA-pieces to be joined together and (usually PCR mediated) synthesis of suitable adapters. LIC cannot thus be regarded as a "non-scaring" sub-cloning method.
References
- Stemmer et al., Single step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides, Gene 1614 (1995) 49-53, doi:10.1016/0378-1119(95)00511-4
- Smith et al., Generating a synthetic genome by whole genome assembly: [var phi]X174 bacteriophage from synthetic oligonucleotides, PNAS, 2003 100(26): 15440–15445.
- ↑ Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. (2009). "Enzymatic assembly of DNA molecules up to several hundred kilobases". Nature Methods 6 (5): 343–345. doi:10.1038/nmeth.1318. PMID 19363495.
- ↑ Gibson Assembly video