Phytochrome
| Phytochrome | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystal Structure of Phytochrome.[1] | |||||||||
| Identifiers | |||||||||
| Symbol | Phytochrome | ||||||||
| Pfam | PF00360 | ||||||||
| InterPro | IPR013515 | ||||||||
| |||||||||
Phytochrome is a photoreceptor, a pigment that plants, and some bacteria and fungi, use to detect light. It is sensitive to light in the red and far-red region of the visible spectrum. Many flowering plants use it to regulate the time of flowering based on the length of day and night (photoperiodism) and to set circadian rhythms. It also regulates other responses including the germination of seeds (photoblasty), elongation of seedlings, the size, shape and number of leaves, the synthesis of chlorophyll, and the straightening of the epicotyl or hypocotyl hook of dicot seedlings. It is found in the leaves of most plants.
Biochemically, phytochrome is a protein with a bilin chromophore.
Phytochrome has been found in most plants including all higher plants; very similar molecules have been found in several bacteria. A fragment of a bacterial phytochrome now has a solved three-dimensional protein structure.
Other plant photoreceptors include cryptochromes, phototropins, and UVR8, which are sensitive to light in the blue and ultra-violet regions of the spectrum.
Structure
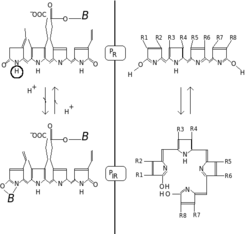
Phytochrome consists of two identical chains (A and B). Each chain has a PAS domain and GAF domain. The PAS domain serves as a signal sensor and the GAF domain is responsible for binding to cGMP and also senses light signals. Together, these subunits form the phytochrome region, which regulates physiological changes in plants to changes in red and far red light conditions. In plants, red light changes phytochrome to its biologically active form, while far red light changes the protein to its biologically inactive form.
Isoforms or states
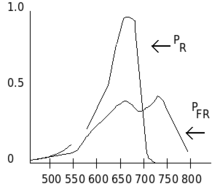
Phytochromes are characterised by a red/far-red photochromicity. Photochromic pigments change their "colour" (spectral absorbance properties) upon light absorption. In the case of phytochrome the ground state is Pr, the r indicating that it absorbs red light particularly strongly. The absorbance maximum is a sharp peak 650–670 nm, so concentrated phytochrome solutions look turquoise-blue to the human eye. But once a red photon has been absorbed, the pigment undergoes a rapid conformational change to form the Pfr state. Here fr indicates that now not red but far-red (also called "near infra-red"; 705–740 nm) is preferentially absorbed. This shift in absorbance is apparent to the human eye as a slightly more greenish colour. When Pfr absorbs far-red light it is converted back to Pr. Hence, red light makes Pfr, far-red light makes Pr. In plants at least Pfr is the physiologically active or "signalling" state.
Biochemistry
Chemically, phytochrome consists of a chromophore, a single bilin molecule consisting of an open chain of four pyrrole rings, bonded to the protein moiety. It is the chromophore that absorbs light, and as a result changes the conformation of bilin and subsequently that of the attached protein, changing it from one state or isoform to the other.
The phytochrome chromophore is usually phytochromobilin, and is closely related to phycocyanobilin (the chromophore of the phycobiliproteins used by cyanobacteria and red algae to capture light for photosynthesis) and to the bile pigment bilirubin (whose structure is also affected by light exposure, a fact exploited in the phototherapy of jaundiced newborns). The term "bili" in all these names refers to bile. Bilins are derived from the closed tetrapyrrole ring of haem by an oxidative reaction catalysed by haem oxygenase to yield their characteristic open chain. Chlorophyll too is derived from haem (Heme). In contrast to bilins, haem and chlorophyll carry a metal atom in the center of the ring, iron or magnesium, respectively.[4]
The Pfr state passes on a signal to other biological systems in the cell, such as the mechanisms responsible for gene expression. Although this mechanism is almost certainly a biochemical process, it is still the subject of much debate. It is known that although phytochromes are synthesized in the cytosol and the Pr form is localized there, the Pfr form, when generated by light illumination, is translocated to the cell nucleus. This implies a role of phytochrome in controlling gene expression, and many genes are known to be regulated by phytochrome, but the exact mechanism has still to be fully discovered. It has been proposed that phytochrome, in the Pfr form, may act as a kinase, and it has been demonstrated that phytochrome in the Pfr form can interact directly with transcription factors.
Discovery
The phytochrome pigment was discovered by Sterling Hendricks and Harry Borthwick at the USDA-ARS Beltsville Agricultural Research Center in Maryland during a period from the late 1940s to the early 1960s. Using a spectrograph built from borrowed and war-surplus parts, they discovered that red light was very effective for promoting germination or triggering flowering responses. The red light responses were reversible by far-red light, indicating the presence of a photoreversible pigment.
The phytochrome pigment was identified using a spectrophotometer in 1959 by biophysicist Warren Butler and biochemist Harold Siegelman. Butler was also responsible for the name, phytochrome.
In 1983 the laboratories of Peter Quail and Clark Lagarias reported the chemical purification of the intact phytochrome molecule, and in 1985 the first phytochrome gene sequence was published by Howard Hershey and Peter Quail. By 1989, molecular genetics and work with monoclonal antibodies that more than one type of phytochrome existed; for example, the pea plant was shown to have at least two phytochrome types (then called type I (found predominantly in dark-grown seedlings) and type II (predominant in green plants)). It is now known by genome sequencing that Arabidopsis has five phytochrome genes (PHYA - E) but that rice has only three (PHYA - C). While this probably represents the condition in several di- and monocotyledonous plants, many plants are polyploid. Hence maize, for example, has six phytochromes - phyA1, phyA2, phyB1, phyB2, phyC1 and phyC2. While all these phytochromes have significantly different protein components, they all use phytochromobilin as their light-absorbing chromophore. Phytohrome A or phyA is rapidly degraded in the Pfr form - much more so than the other members of the family. In the late 1980s, the Vierstra lab showed that phyA is degraded by the ubiquitin system, the first natural target of the system to be identified in eukaryotes.
In 1996 a gene in the newly sequenced genome of the cyanobacterium Synechocystis was noticed to have a weak similarity to those of plant phytochromes, the first evidence of phytochromes outside the plant kingdom. Jon Hughes in Berlin and Clark Lagarias at UC Davis subsequently showed that this gene indeed encoded a bona fide phytochrome (named Cph1) in the sense that it is a red/far-red reversible chromoprotein. Presumably plant phytochromes are derived from an ancestral cyanobacterial phytochrome, perhaps by gene migration from the chloroplast to the nucleus. Subsequently phytochromes have been found in other prokaryotes including Deinococcus radiodurans and Agrobacterium tumefaciens. In Deinococcus phytochrome regulates the production of light-protective pigments, however in Synechocystis and Agrobacterium the biological function of these pigments is still unknown.
In 2005, the Vierstra and Forest labs at the University of Wisconsin published a three-dimensional structure of the photosensory domain of Deinococcus phytochrome. This breakthrough paper revealed that the protein chain forms a knot - a highly unusual structure for a protein.
Genetic engineering
Around 1989 several laboratories were successful in producing transgenic plants which produced elevated amounts of different phytochromes (overexpression). In all cases the resulting plants had conspicuously short stems and dark green leaves. Harry Smith and co-workers at Leicester University in England showed that by increasing the expression level of phytochrome A (which responds to far-red light), shade avoidance responses can be altered.[5] As a result, plants can expend less energy on growing as tall as possible and have more resources for growing seeds and expanding their root systems. This could have many practical benefits: for example, grass blades that would grow more slowly than regular grass would not require mowing as frequently, or crop plants might transfer more energy to the grain instead of growing taller.
References
- ↑ PDB: 3G6O; Yang X, Kuk J, Moffat K (2009). "Crystal structure of P. aeruginosa bacteriaphytochrome PaBphP photosensory core domain mutant Q188L". Proc. Natl. Acad. Sci. U.S.A. 106: 15639–15644. doi:10.2210/pdb3g6o/pdb. PMID 1972099.
- ↑ Britz SJ, Galston AW.. Physiology of Movements in the Stems of Seedling Pisum sativum L. cv Alaska : III. Phototropism in Relation to Gravitropism, Nutation, and Growth, Plant Physiol. 1983 Feb;71(2):313-318
- ↑ Walker TS, Bailey JL. Two spectrally different forms of the phytochrome chromophore extracted from etiolated oat seedlings. Biochem J. 1968 Apr;107(4):603–605.
- ↑ Mauseth, James D. (2003). Botany : An Introduction to Plant Biology (3rd ed.). Sudbury, MA: Jones and Bartlett Learning. pp. 422–427. ISBN 0-7637-2134-4.
- ↑ Robson, P. R. H., McCormac, A. C., Irvine, A. S. & Smith, H. Genetic engineering of harvest index in tobacco through overexpression of a phytochrome gene. Nature Biotechnol. 14, 995–998 (1996).
6. ^Gururani, Mayank Anand, Markkandan Ganesan, and Pill-Soon Song. "Photo-biotechnology as a tool to improve agronomic traits in crops." Biotechnology Advances (2014).
Other sources
- Hua Lia, Junrui Zhangb, Richard D. Vierstra, and Huilin Lia Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy PNAS June 1, 2010, doi:10.1073/pnas.1001908107
- http://www.ars.usda.gov/is/timeline/light.htm
- http://www.mobot.org/jwcross/duckweed/phytochrome.htm#tetrapyrrole
- http://ucce.ucdavis.edu/files/filelibrary/616/17562.htm
- Terry and Gerry Audesirk. Biology: Life on Earth.
- Linda C Sage. A pigment of the imagination: a history of phytochrome research. Academic Press 1992. ISBN 0-12-614445-1
|