Phospholipid-derived fatty acids
Phospholipid-derived fatty acids (PLFA) are widely used in microbial ecology as chemotaxonomic markers of bacteria and other organisms. Phospholipids are the primary lipids composing cellular membranes. Phospholipids can be saponified, which releases the fatty acids contained in their diglyceride tail. Once the phospholipids of an unknown sample are saponified, the composition of the resulting PLFA can be compared to the PLFA of known organisms to determine the identity of the sample organism. PLFA analysis may be combined with other techniques, such as stable isotope probing to determine which microbes are metabolically active in a sample. PLFA analysis was pioneered by D.C. White, MD, PhD,[1] at the University of Tennessee, in the early to mid 1980s.

Phospholipid fatty acid (PLFA) analysis
Phospholipid fatty acids (PLFA) are an essential structural component of all microbial cellular membranes. PLFA analysis is a technique widely used for estimation of the total biomass and to observe broad changes in the community composition of the living microbiota of soil and aqueous environments. There has been a surge of interest in PLFAs in recent years, evident from the large increase in peer-reviewed journal references on the subject.[2] However, there is increasing concern that some researchers are assigning PLFAs to specific microbial classes when in fact those PLFAs are present in a broad range of life forms.[2] Phospholipids can occur in many biological classes (such as in plant roots, fungi, as well as in soil bacteria), so care has to be taken in over-assigning PLFA biomarkers to the wrong class. Even though phospholipids occur in many different life forms, the fatty acid side chains between differing life forms can be quite unique. Polyunsaturated fatty acids (e.g. 18:3 ω3c) are found in plants, algae and cyanobacteria, but are often not present in bacteria. Monounsaturated fatty acids (particularly at the omega-7 position), odd-chain saturated fatty acids (e.g. 15:0), branched-chain fatty acids (mainly iso/anetiso and 10-methyl) and cyclopropane fatty acids (e.g. 19:0 cyclo ω7c) are mostly synthesized by bacteria. The monounsaturated fatty acid, 16:1 ω5c, is mostly synthesized by Arbuscular mycorrhizal fungi (AMF) and the polyunsaturated fatty acid, 18:2 ω6c (Linoleic acid), is mostly synthesized by Ectomycorrhizal fungi.
The basic premise is that as individual organisms (especially bacteria and fungi) die, phospholipids are rapidly degraded and the remaining phospholipid content of the sample is assumed to be from living organisms. As the phospholipids of different groups of bacteria and fungi contain a variety of somewhat unique fatty acids, they can serve as useful biomarkers for such groups. PLFA profiles and composition can be determined by purifying the phospholipids and then cleaving the fatty acids for further analysis. Knowledge of the composition and metabolic activity of the microbiota in soils, water and waste materials is useful in optimizing crop production, in bioremediation and in understanding microbial ecosystems. Soil microbial community analysis by PLFA has been a widely used technique due to the sensitive, reproducible measurement of the dominant portions of the soil microbiota and the fact that PLFA does not require cultivation of the organisms.[3] Sampling of soil populations by culturing has proven not cost effective and results in biased results due to the differing ease of culturing of some organisms. The main drawback of PLFA has been that the extraction time is very long and cumbersome. A new 96-well plate PLFA extraction procedure has been developed which represents a 4-to-5 fold increase in throughput over traditional PLFA extraction methods. This new method, coupled to new software tools for analyzing the PLFA data, will be useful to laboratories performing large numbers of PLFA analyses, or for laboratories wanting to begin PLFA research.[4]

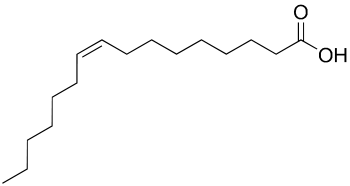
Some Phospholipid fatty acid biomarkers (common case)
- Saturated fatty acids (SAFA)
- • 15:0 (Pentadecanoic acid) – Bacteria
- • Other straight chain (e.g. 16:0, Palmitic acid) – Prokaryotes and Eukaryotes
- • iso-branched (e.g. 17:0 iso, 15-Methylpalmitic acid) – Gram-positive bacteria
- • anteiso-branched (e.g. 17:0 anteiso, 14-Methylpalmitic acid) – Gram-positive bacteria
- • 10-methyl branched (e.g. 19:0 10-methyl, Tuberculostearic acid) – Actinomycetes
- Monounsaturated fatty acids (MUFA)
- • 16:1 ω5c (Hexadecenoic acid) - Arbuscular mycorrhizal fungi (AMF)
- • Omega-5 and 7 positions (e.g. 16:1 ω7c, Palmitoleic acid) – Gram-negative bacteria
- • 16:1 ω8c (8-Hexadecenoic acid) - Methane-oxidizing bacteria Type I
- • 18:1 ω8c (10-Octadecenoic acid) - Methane-oxidizing bacteria Type II
- • Omega-9 position (e.g. 16:1 ω9c, cis-7-Palmitoleic acid) - Ectomycorrhizal fungi & Gram-positive bacteria
- Polyunsaturated fatty acids (PUFA)
- • 18:2 ω6c, (Linoleic acid) – Ectomycorrhizal fungi
- • 20:2 ω6c, 20:3 ω6c, 20:4 ω6c - Protozoa
- • Other PUFAs - Eukaryotes
- Cyclopropane fatty acids (e.g. 19:0 cyclo ω7c) – Gram-negative bacteria
- Dimethyl acetal (e.g. 16:0 DMA, Hexadecanal dimethyl acetal) – Anaerobic bacteria
Background of PLFA analysis
Early studies of the living soil microbial communities were largely based on attempts at culturing bacteria and fungi of soil. However, due to difficulty in culturing many of the organisms, the differential growth rates of the organisms, and labor involved, this proved to be not satisfactory. A 1965 article proposed using molecules produced by the organisms as biomarkers for the microbial communities.[5] In the following two decades, rapid progress was made in development of gas chromatographs (GC) and of fused silica capillary columns for the GC instruments, enabling better analysis of biological materials, including fatty acid methyl esters (FAMEs). PLFA analysis can be used for microbial community structure and activity through the use of “signature” fatty acids.[6] The basic concept is that the phospholipid content represents living organisms as these compounds are rapidly decomposed in aerobic mixed communities and that some of the neutral lipid components such as the lipopolysaccharides of Gram-negative bacteria do not reflect organisms alive at the time of sampling.
PLFA sample preparation
Although the method of sample collection is different for soil, water samples, etc., the extraction-derivatization is generally similar to the following protocol from an article on soil microbial communities.[7] The lipids were extracted from the dried soil sample by use of a chloroform-methanol-phosphate buffer mixture by use of a brief sonication followed by shaking for 2 hours and centrifugation to pellet the soil material. The liquid above the soil had additional chloroform and water added to cause separation of the lipid-containing chloroform from the buffer/methanol phase. The lipids were fractionated on a solid-phase extraction column and the neutral lipids, free fatty acids and other materials discarded and the phospholipid phase then dried, prior esterification to form the fatty acid methyl esters (FAMEs)[7] to make them suitable for analysis.
Analysis of FAMEs
Analysis by gas chromatography (GC) is the method of choice for FAME analysis. The GC is coupled with either a mass spectrometer detector (MSD) or a flame ionization detector (FID). The GC-MSD system is more expensive to purchase and to maintain as well as requiring considerable skill in operation. Identification of fatty acids using the GC-FID system is dependent on comparison of retention times of the compounds in comparison to purchased standards of fatty acid esters. A commercially available, fatty-acid based microbial identification system (using GC-FID), which reproducibly names and quantitates the FAMEs, has been widely adopted for PLFA analysis.[8]
The PLFA components of soil microbiota
Actinomycetes are Gram-positive bacteria that are some of the most common bacteria in soil, freshwater and marine environments. Actinomycetes are active in decomposition of organic matter and give rise to the rich “earthy” smell of freshly tilled soils. This group of bacteria produce distinctive biomarker fatty acids having a methyl branch at the 10th carbon, such as 16:0 10-methyl and 18:0 10-methyl.[9] Some common species of soil actinomycetes include Rhodococcus, Nocardia, Corynebacterium and Streptomyces.
Gram-positive bacteria include aerobic Bacillus species especially those related to B. cereus and to B. subtilis. These bacteria are common in the bulk soil and increase in numbers in the rhizosphere. The PLFA profiles of these Gram-positive species have high percentages of biomarker branched-chain fatty acids such as 15:0 iso and 15:0 anteiso. Thus, the sum of the iso and anteiso fatty acids in a PLFA analysis may provide an estimate of the abundance of the Gram-positive bacteria (other than actinomycetes) in the sample.
Gram-negative bacteria are a major component of the plant rhizosphere and improve plant growth by increasing solubility of phosphate, producing ionophore compounds that increase uptake of iron or other minerals and may produce antifungal compounds.[10] Gram-negative bacteria produce high levels of monounsaturated fatty acids (e.g. 16:1 omega-7 and 18:1 omega-9) during active metabolism but convert much of the unsaturated fatty acid composition to cyclopropane fatty acids (e.g. 17:0 cyclopropane and 19:0 cyclopropane) when metabolism and cell division slow due to shortage of nutrition or other stress. Thus, in PLFA analysis, the sum of monounsaturated and cyclopropane fatty acids may provide an estimate of the abundance of Gram-negative bacteria. A high ratio of cyclopropane to monounsaturated fatty acid indicates stress conditions.[3]
Anaerobic bacteria in agriculture are primarily a factor in soils of low oxygen levels such as occur in greater depths or of wet conditions such as in rice paddies. Using PLFA analysis in early sampling, the bacteria- archaea consortia in rice paddy soil was about 44% aerobic bacteria, 32% facultatively anaerobic- bacteria and 24% archaea. Under longer term flooding, the levels were 27%, 36% and 37% respectively and with total biomass being significantly lower.[11] Dimethyl acetals (DMA) formed during derivatization are considered to be biomarkers of anaerobic bacteria.[12]
Archaea are universally distributed in soils and have been shown to control nitrification in acidic conditions [13] and to contribute to ammonia oxidation in agricultural and forest soils.[14] However, as the phospholipids of archaea are not ester linked as in bacteria, but are ether linked, they are not significantly present in routine PLFA sample preparation which is designed to cleave ester-linked fatty acids.
Arbuscular mycorrhizae fungi (AMF) penetrate the walls of cortical cells of about 80% of all vascular plant families, generating a symbiotic relationship. The fungi form membrane structures adjacent to the plant cell membrane allowing exchange of phosphorus, nitrogen compounds and minerals from the fungus and the plant provides the fungus primarily with photosynthesis-derived sugars. As the AMF are obligate symbiotic fungi, they are not free-living in the soil. The AMF hyphae in the root form lipid materials which then are transported to the hyphae that extend into the soil from the root and thus may occur in a soil sample.[15] Vesicles are lipid storage organs of AMF and these and the hyphae in the soil contain the fatty acids 18:2 w6c (often used as an indicator of fungal content of the PLFA analysis) as well as containing the fatty acid 16:1 w5c which has been recommended as a biomarker for AMF.[16]
Applications of PLFA analysis
Sampling of agricultural soils for analysis of chemical composition (e.g. pH, N, P, K, Ca, Mg, etc.) has long been practiced in crop production and while there has been recognition of the importance of the soil microbiota, tools for studying the microbiota have been developed relatively recently.
High value vegetable crops
Many high-value vegetable crops easily justify soil testing both for chemical content and the soil microbiota.[17] Conventional, low-input and organic farming systems showed a rapid response of the soil microbial communities to wet/dry cycles and that increases in bacterial cyclopropyl fatty acids were useful to detect periods of stress.[18] Lines of transgenic corn (maize) expressing Bacillus thuringiensis endotoxins were found to have small effect on soil microbial communities when compared by PLFA analysis to their non-transgenic isolines.[7] Successful exotic invasive plant species can have profound effects on the microbial communities of the soil [19] perhaps thus improving their competitiveness. Grassland restoration practices of tillage, weeding and herbicide use showed an impact on microbial communities of the upper soil but very small changes on the microbiota of lower soil layers and that after 4 years of recovery the communities were very similar to untreated plots.[20]
Bioremediation
Bioremediation has been studied using PLFA analysis of soil microbiota from sites contaminated by diesel oil,[21] crude oil,[22] explosives,[23] olive mill waste,[24] pentachlorophenol,[25] coal tar [26] and PCBs.[27] There are reports of the effects on PLFAs of heavy metals on arbuscular fungi [28] and on bacteria,[9] of polycyclic aromatic hydrocarbons on rice paddy bacteria [29] and of methylene chloride on bacteria.[30]
Phytoplankton
Phytoplankton (eukaryotic algae) are microscopic photosynthesizing plants that inhabit the sunlit layers of oceans and bodies of freshwater. As the primary source of elaborated carbon compounds, they are vital to the aquatic food web. Phytoplankton produce considerable amounts of polyunsaturated fatty acids (PUFA), including Eicosapentaenoic acid (EPA, 20:5 w3c), with microalgae being the origin of omega-3 fatty acids in fish oil.[31] The diverse taxonomic groups in algae vary in abundance dependent on environmental conditions such as temperature, salinity, sunlight, and nutrient availability. The PLFA biomarker compositions were found to enable determination of the prevalence of the major groups in several marine environments.[32] In a study of reservoir sedimentary desposits, an assumption was made that the community PUFA content constituted ca. 50% of the total microeukaryotic PLFAs.[33] It was also assumed that “The ratio of omega-3 to omega-6 fatty acids describes the relative contribution of phototrophic to heterotrophic members of the microeukaryotic community…[33]”.
Aquatic environments
In contrast to the considerable microbial diversity in soils, free-living microbes distributed by marine currents and exposed to algal exudates exhibit global distributions for a few dominant microbial groups of relatively few species.[34] Streambed sediments displayed a variation in microbial community structure (as measured by PLFA) related to the forest environment and geographic location of the stream, with much of the variation determined by use of the algal biomarker fatty acid 18:3 w3.[35] By PLFA analysis, considerable spatial and seasonal variations were determined in a freshwater reservoir sedimentary microbial community.[33]
Forestry
Coniferous forests are dependent on available nutrients in soil rather than agricultural fertilizers and thus are routinely colonized by symbiotic mycorrhizal fungi. The mycorrhizae may be ectomycorrhizae (EMF) and/or arbuscular (AMF) in type in the forest.[36] The amount of total PLFA in soil provides an estimate of the total soil fungi (not including AMF). The AMF can be estimated by the amount of 16:1 w5c fatty acid in the PLFA.[36] Water stress was indicated by an increase in [PLFA ratios of saturated, monounsaturated and (cyclo 17:0 + cyclo 19:0) / (16:1 w7c + 18:1 w7c)] in a Douglas fir forest.[37] Boreal forests with low soil pH values had elevated EM PLFAs and raising the pH of the soil increased bacterial PLFAs.[38] The introduction of photosynthates through tree roots is a major source of carbon for soil microbiota and influences the composition of fungal and bacterial communities.[39] Forest areas without tree roots had less fungal biomarkers and more actinobacterial biomarkers than areas with tree roots.[40] Addition of nitrogen fertilizer to an oak forest reduced the ectomycorrhizal fungal content of the soil microbiota.[41]
Composting
Composting of organic materials is the microbial degradation of heterogeneous organic material under moist, self-heating, aerobic conditions. Initially, activity by mesophilic organisms leads to a rapid rise in temperature, followed by thermophilic organisms dominating the degradation process leading to a cooling period in which mesophilic bacteria again dominate populations. A commercial FAME extraction protocol developed for identification of bacteria, a mild alkaline methanolysis protocol and PLFA-extraction/derivatization were compared for effectiveness.[42] The PLFA protocol gave the most detailed information about community succession, however, the other two protocols were much simpler and appeared suitable for analysis of microbial FAME profiles in compost.[42]
Wastewater treatment
Activated sludge technology is the most widely used method for wastewater treatment. Complex microbial communities in activated sludge processes are needed for the stable removal efficiency of organic pollutants. PLFA analysis can be used to monitor the microbial community composition of activated sludge reactors, which microbial groups are predominant, and the efficiency of such systems.[43][44]
References
- L Zelles, QY Bai, R Rackwitz, D Chadwick, F Beese (1995). "Determination of phospholipid- and lipopolysaccharide-derived fatty acids as an estimate of microbial biomass and community structures in soils". Biology and Fertility of Soils. Volume 19, Numbers 2-3: 115-123.
- N Dijkman, J Kromkamp (2006). "Phospholipid-derived fatty acids as chemotaxonomic markers for phytoplankton: application for inferring phytoplankton composition". Marine Ecology Progress Series. Vol. 324: 113–125.
- J Neufeld, M Dumont, J Vohra, J Murrell (2007). "Methodological Considerations for the Use of Stable Isotope Probing in Microbial Ecology". Microbial Ecology. Volume 53, Number 3: 435-442.
- ↑ Morris, B.E.L., et al., On the contributions of David Cleaveland White, MD, PhD to microbial ecology: celebrating the life of a pioneer. ISME J, 2008. 2: p. 797-804
- 1 2 Frostegard, A., A. Tunlid, and E. Baath, Use and misuse of PLFA measurements in soil. Soil Biol & Biochem, 2011. 43(8): p 1621-25
- 1 2 Kaur, A., et al., Phospholipid fatty acid - A bioindicator of environmental monitoring and assessment in soil ecosystem. Current Science, 2005. 89(7): p. 1103-1112
- ↑ Buyer, J.S., and M. Sasser, High throughput fatty acid analysis of soils. App Soil Ecol, 61(2012): p 127-130 | http://dx.doi.org/10.1016/j.apsoil.2012.06.005
- ↑ Zuckerkandl, E. and L. Pauling, Molecules as documents of evolutionary history. J Theor Biol, 1965. 8(2): p. 357-66
- ↑ White, D.C., Analysis of microorganisms in terms of quantity and activity in natural environments., in Microbes in Their Natural Environment, J.H.W. Slater, R. Wimpenny, J.W.T., Editor 1983 Cambridge University Press
- 1 2 3 Blackwood, C.B. and J.S. Buyer, Soil microbial communities associated with Bt and non-Bt corn in three soils. J Environ Qual, 2004. 33(3): p. 832-6
- ↑ Piotrowska-Seget, Z. and A. Mrozik, Signature lipid biomarker (SLB) analysis in determining changes in community structure of soil microorganisms. Polish J Environ Stud, 2003. 12(6): 669-75
- 1 2 Frostegard, A., A. Tunlid, and E. Baath, Phospholipid Fatty Acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl Environ Microbiol, 1993. 59(11): p. 3605-17.
- ↑ Pandey, A., et al., Characterization of a phosphate solubilizing and antagonistic strain of Pseudomonas putida (B0) isolated from a sub-alpine location in the Indian Central Himalaya. Curr Microbiol, 2006. 53(2): p. 102-7
- ↑ Bai, Q., A. Gattinger, and L. Zelles, Characterization of Microbial Consortia in Paddy Rice Soil by Phospholipid Analysis. Microb Ecol, 2000. 39(4): p. 273-281
- ↑ Zelles, L., Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol Fertil Soils, 1999. 29(2): p. 111-129
- ↑ Gubry-Rangin, C., G.W. Nicol, and J.I. Prosser, Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol, 2010. 74(3): p. 566-74
- ↑ Szukics, U., et al., Rapid and dissimilar response of ammonia oxidizing archaea and bacteria to nitrogen and water amendment in two temperate forest soils. Microbiol Res, 2011
- ↑ Pfeffer, P.E., et al., Carbon uptake and the metabolism and transport of lipids in an arbuscular mycorrhiza. Plant Physiol, 1999. 120(2): p. 587-98.
- ↑ van Aarle, I.M. and P.A. Olsson, Fungal lipid accumulation and development of mycelial structures by two arbuscular mycorrhizal fungi. Appl Environ Microbiol, 2003. 69(11): p. 6762-7
- ↑ Buyer, J.S., et al., Factors affecting soil microbial community structure in tomato cropping systems. Soil Biol & Biochem, 2010. 42(5): p. 831-41
- ↑ Lundquist, E.J., et al., Rapid response of soil microbial communities from conventional, low input, and organic farming systems to a wet/dry cycle. Soil Biol & Biochem, 1999. 31(12): p. 1661-75
- ↑ Kourtev, P.S., J.G. Ehrenfeld, and M. Haggblom, Exotic plant species alter the microbial community structure and function in the soil. Ecology, 2002. 83(11): p. 3152-66
- ↑ Potthoff, M., et al., Soil microbial community composition as affected by restoration practices in California grassland. Soil Biol & Biochem, 2006. 38(7): p. 1851-60
- ↑ Ringelberg, D., et al., Utility of lipid biomarkers in support of bioremediation efforts at army sites. J Microbiol Methods, 2008. 74(1): p. 17-25
- ↑ MacNaughton, S.J., et al., Microbial population changes during bioremediation of an experimental oil spill. Appl Environ Microbiol, 1999. 65(8): p. 3566-74
- ↑ Fuller, M.E. and J.F. Manning, Jr., Microbiological changes during bioremediation of explosives-contaminated soils in laboratory and pilot-scale bioslurry reactors. Bioresour Technol, 2004. 91(2): p. 123-33
- ↑ Morillo, J.A., et al., Molecular microbial and chemical investigation of the bioremediation of two-phase olive mill waste using laboratory-scale bioreactors. Appl Microbiol Biotechnol, 2008. 79(2): p. 309-17
- ↑ Hansen, L.D., et al., Extended bioremediation of PAH/PCP contaminated soils from the POPILE wood treatment facility. Chemosphere, 2004. 54(10): p. 1481-93
- ↑ Antizar-Ladislao, B., et al., The influence of different temperature programmes on the bioremediation of polycyclic aromatic hydrocarbons (PAHs) in a coal-tar contaminated soil by in-vessel composting. J Hazard Mater, 2007. 144(1-2): p. 340-7
- ↑ Slater, H., T. Gouin, and M.B. Leigh, Assessing the potential for rhizoremediation of PCB contaminated soils in northern regions using native tree species. Chemosphere, 2011. 84(2): p. 199-206
- ↑ Hassan Sel, D., et al., Molecular biodiversity of arbuscular mycorrhizal fungi in trace metal-polluted soils. Mol Ecol, 2011. 20(16): p. 3469-83
- ↑ Su, Y.H. and X.Y. Yang, Interactions between selected PAHs and the microbial community in rhizosphere of a paddy soil. Sci Total Environ, 2009. 407(3): p. 1027-34
- ↑ Schwartz, E., S.V. Trinh, and K.M. Scow, Impact of methylene chloride on microorganisms and phenanthrene mineralization in soil. J Environ Qual, 2002. 31(1): p. 144-9.29
- ↑ Wen, Z.Y. and F. Chen, Heterotrophic production of eicosapentaenoic acid by microalgae. Biotechnol Adv, 2003. 21(4): p. 273-94
- ↑ Djikman, N.A., et al., Phospholipid-derived fatty acids as chemotaxonomic markers for phytoplankton: application for inferring phytoplankton composition. Marine Ecol Progress, 2006. 324: p. 113-25
- 1 2 3 Smoot, J.C. and R.H. Findlay, Spatial and Seasonal Variation in a Reservoir Sedimentary Microbial Community as Determined by Phospholipid Analysis. Microb Ecol, 2001. 42(3): p. 350-358
- ↑ Hagstrom, A., et al., Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl Environ Microbiol, 2002. 68(7): p. 3628-33
- ↑ Findlay, R.H., et al., Biome-level biogeography of streambed microbiota. Appl Environ Microbiol, 2008. 74(10): p. 3014-21
- 1 2 Nilsson, L.O., et al., Growth and biomass of mycorrhizal mycelia in coniferous forests along short natural nutrient gradients. New Phytol, 2005. 165(2): p. 613-22
- ↑ Moore-Kucera, J. and R.P. Dick, PLFA profiling of microbial community structure and seasonal shifts in soils of a Douglas-fir chronosequence. Microb Ecol, 2008. 55(3): p. 500-11
- ↑ Hogberg, M.N., P. Hogberg, and D.D. Myrold, Is microbial community composition in boreal forest soils determined by pH, C-to-N ratio, the trees, or all three? Oecologia, 2007. 150(4): p. 590-601
- ↑ Yarwood, S.A., D.D. Myrold, and M.N. Hogberg, Termination of below ground C allocation by trees alters soil fungal and bacterial communities in a boreal forest. FEMS Microbiol Ecol, 2009. 70(1): p. 151-62
- ↑ Brant, J.B., D.D. Myrold, and E.W. Sulzman, Root controls on soil microbial community structure in forest soils. Oecologia, 2006. 148(4): p. 650-9
- ↑ Nilsson, L.O., et al., Growth of ectomycorrhizal mycelia and composition of soil microbial communities in oak forest soils along a nitrogen deposition gradient. Oecologia, 2007. 153(2): p. 375-84
- 1 2 Steger, K., et al., Comparison of signature lipid methods to determine microbial community structure in compost. J Microbiol Methods, 2003. 55(2): p. 371-82
- ↑ Yi, T., et al., Structure and dynamics of microbial community in full-scale activated sludge reactors. J Ind Microbio Biotechnol, 2011. In Press
- ↑ Chang, J-J., et al., Effect of intermittent aeration on the microbial community structure of activated sludge in a submerged membrane bioreactor. Water Environ J, 2011. 25(2): p. 214-18