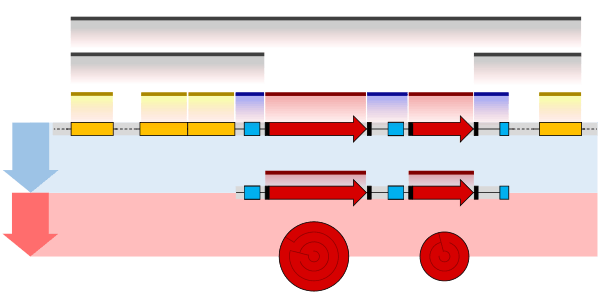
Operon
In genetics, an operon is a functioning unit of genomic DNA containing a cluster of genes under the control of a single promoter.[1] The genes are transcribed together into an mRNA strand and either translated together in the cytoplasm, or undergo trans-splicing to create monocistronic mRNAs that are translated separately, i.e. several strands of mRNA that each encode a single gene product. The result of this is that the genes contained in the operon are either expressed together or not at all. Several genes must be co-transcribed to define an operon.[2]
Originally, operons were thought to exist solely in prokaryotes, but since the discovery of the first operons in eukaryotes in the early 1990s,[3][4] more evidence has arisen to suggest they are more common than previously assumed.[5] In general, expression of prokaryotic operons leads to the generation of polycistronic mRNAs, while eukaryotic operons lead to monocistronic mRNAs.
Operons are also found in viruses such as bacteriophages.[6][7] For example, T7 phages have two operons. The first operon codes for various products, including a special T7 RNA polymerase which can bind to and transcribe the second operon. The second operon includes a lysis gene meant to cause the host cell to burst.[8]
History
The term "operon" was first proposed in a short paper in the Proceedings of the French Academy of Science in 1960.[9] From this paper, the so-called general theory of the operon was developed. This theory suggested that in all cases, genes within an operon are negatively controlled by a repressor acting at a single operator located before the first gene. Later, it was discovered that genes could be positively regulated and also regulated at steps that follow transcription initiation. Therefore, it is not possible to talk of a general regulatory mechanism, because different operons have different mechanisms. Today, the operon is simply defined as a cluster of genes transcribed into a single mRNA molecule. Nevertheless, the development of the concept is considered a landmark event in the history of molecular biology. The first operon to be described was the lac operon in E. coli.[9] The 1965 Nobel Prize in Physiology and Medicine was awarded to François Jacob, André Michel Lwoff and Jacques Monod for their discoveries concerning the operon and virus synthesis.
Overview
|
|
Operons occur primarily in prokaryotes but also in some eukaryotes, including nematodes such as C. elegans and the fly, Drosophila melanogaster. rRNA genes often exist in operons that have been found in a range of eukaryotes including chordates. An operon is made up of several structural genes arranged under a common promoter and regulated by a common operator. It is defined as a set of adjacent structural genes, plus the adjacent regulatory signals that affect transcription of the structural genes.5[10] The regulators of a given operon, including repressors, corepressors, and activators, are not necessarily coded for by that operon. The location and condition of the regulators, promoter, operator and structural DNA sequences can determine the effects of common mutations.
Operons are related to regulons, stimulons and modulons; whereas operons contain a set of genes regulated by the same operator, regulons contain a set of genes under regulation by a single regulatory protein, and stimulons contain a set of genes under regulation by a single cell stimulus. According to its authors, the term "operon" means "to operate".[11]
As a unit of transcription
An operon contains one or more structural genes which are generally transcribed into one polycistronic mRNA (a single mRNA molecule that codes for more than one protein). However, the definition of an operon does not require the mRNA to be polycistronic, though in practice, it usually is.[12] Upstream of the structural genes lies a promoter sequence which provides a site for RNA polymerase to bind and initiate transcription. Close to the promoter lies a section of DNA called an operator.
Operons versus clustering of prokaryotic genes
All the structural genes of an operon are turned ON or OFF together, due to a single promoter and operator upstream to them, but sometimes more control over the gene expression is needed. To achieve this aspect, some bacterial genes are located near together, but there is a specific promoter for each of them; this is called gene clustering. Usually these genes encode proteins which will work together in the same pathway, such as a metabolic pathway. Gene clustering helps a prokaryotic cell to produce metabolic enzymes in a correct order.
General structure of an operon

An operon is made up of 3 basic DNA components:
- Promoter – a nucleotide sequence that enables a gene to be transcribed. The promoter is recognized by RNA polymerase, which then initiates transcription. In RNA synthesis, promoters indicate which genes should be used for messenger RNA creation – and, by extension, control which proteins the cell produces.
- Operator – a segment of DNA that a repressor binds to. It is classically defined in the lac operon as a segment between the promoter and the genes of the operon.[13] In the case of a repressor, the repressor protein physically obstructs the RNA polymerase from transcribing the genes.
- Structural genes – the genes that are co-regulated by the operon.
Not always included within the operon, but important in its function is a regulatory gene, a constantly expressed gene which codes for repressor proteins. The regulatory gene does not need to be in, adjacent to, or even near the operon to control it.[14]
Regulation
Control of an operon is a type of gene regulation that enables organisms to regulate the expression of various genes depending on environmental conditions. Operon regulation can be either negative or positive by induction or repression.[13]
Negative control involves the binding of a repressor to the operator to prevent transcription.
- In negative inducible operons, a regulatory repressor protein is normally bound to the operator, which prevents the transcription of the genes on the operon. If an inducer molecule is present, it binds to the repressor and changes its conformation so that it is unable to bind to the operator. This allows for expression of the operon. The lac operon is a negatively controlled inducible operon, where the inducer molecule is allolactose.
- In negative repressible operons, transcription of the operon normally takes place. Repressor proteins are produced by a regulator gene, but they are unable to bind to the operator in their normal conformation. However, certain molecules called corepressors are bound by the repressor protein, causing a conformational change to the active site. The activated repressor protein binds to the operator and prevents transcription. The trp operon, involved in the synthesis of tryptophan (which itself acts as the corepressor), is a negatively controlled repressible operon.
Operons can also be positively controlled. With positive control, an activator protein stimulates transcription by binding to DNA (usually at a site other than the operator).
- In positive inducible operons, activator proteins are normally unable to bind to the pertinent DNA. When an inducer is bound by the activator protein, it undergoes a change in conformation so that it can bind to the DNA and activate transcription.
- In positive repressible operons, the activator proteins are normally bound to the pertinent DNA segment. However, when an inhibitor is bound by the activator, it is prevented from binding the DNA. This stops activation and transcription of the system.
The lac operon
The lac operon of the model bacterium Escherichia coli was the first operon to be discovered and provides a typical example of operon function. It consists of three adjacent structural genes, a promoter, a terminator, and an operator. The lac operon is regulated by several factors including the availability of glucose and lactose. This is an example of the derepressible (from above: negative inducible) model.
The trp operon
Discovered in 1953 by Jacques Monod and colleagues, the trp operon in E. coli was the first repressible operon to be discovered. While the lac operon can be activated by a chemical (allolactose), the tryptophan (Trp) operon is inhibited by a chemical (tryptophan). This operon contains five structural genes: trp E, trp D, trp C, trp B, and trp A, which encodes tryptophan synthetase. It also contains a promoter which binds to RNA polymerase and an operator which blocks transcription when bound to the protein synthesized by the repressor gene (trp R) that binds to the operator. In the lac operon, lactose binds to the repressor protein and prevents it from repressing gene transcription, while in the trp operon, tryptophan binds to the repressor protein and enables it to repress gene transcription. Also unlike the lac operon, the trp operon contains a leader peptide and an attenuator sequence which allows for graded regulation.[15] This is an example of the corepressible model.
Predicting the number and organization of operons
The number and organization of operons has been studied most critically in E. coli. As a result, predictions can be made based on an organism's genomic sequence.
One prediction method uses the intergenic distance between reading frames as a primary predictor of the number of operons in the genome. The separation merely changes the frame and guarantees that the read through is efficient. Longer stretches exist where operons start and stop, often up to 40–50 bases.[16]
An alternative method to predict operons is based on finding gene clusters where gene order and orientation is conserved in two or more genomes.[17]
Operon prediction is even more accurate if the functional class of the molecules is considered. Bacteria have clustered their reading frames into units, sequestered by co-involvement in protein complexes, common pathways, or shared substrates and transporters. Thus, accurate prediction would involve all of these data, a difficult task indeed.
Pascale Cossart's laboratory was the first to experimentally identify all operons of a microorganism, Listeria monocytogenes. The 517 polycistronic operons are listed in a 2009 study describing the global changes in transcription that occur in L. monocytogenes under different conditions.[18]
See also
References
- ↑ Sadava, David; et al. (2009). Life: The Science of Biology (9th ed.). Macmillan. p. 349. ISBN 978-1-4292-1962-4.
- ↑ Lodish, Harvey; Zipursky, Lawrence; Matsudaira, Paul; Baltimore, David; Darnel, James (2000). "Chapter 9: Molecular Definition of a Gene". Molecular Cell Biology. W. H. Freeman. ISBN 978-0-7167-3136-8.
- ↑ Spieth, J.; Brooke, G.; Kuersten, S.; Lea, K.; Blumenthal, T. (1993). "Operons in C. Elegans: polycistronic mRNA precursors are processed by trans-splicing of SL2 to downstream coding regions". Cell 73 (3): 521–532. doi:10.1016/0092-8674(93)90139-H. PMID 8098272.
- ↑ Brogna, S.; Ashburner, M. (1997). "The Adh-related gene of Drosophila melanogaster is expressed as a functional dicistronic messenger RNA: Multigenic transcription in higher organisms". The EMBO Journal 16 (8): 2023–2031. doi:10.1093/emboj/16.8.2023. PMC 1169805. PMID 9155028.
- ↑ Blumenthal, T. (2004). "Operons in eukaryotes". Briefings in Functional Genomics and Proteomics 3 (3): 199–211. doi:10.1093/bfgp/3.3.199. PMID 15642184.
- ↑ "Definition of Operon". Medical Dictionary. MedicineNet.com. Retrieved 30 December 2012.
- ↑ "Displacements of Prohead Protease Genes in the Late Operons of Double-Stranded-DNA Bacteriophages". Journal of Bacteriology. 1 March 2004. Retrieved 30 December 2012.
- ↑ "Bacteriophage Use Operons". Prokaryotic Gene Control. Dartmouth College. Retrieved 30 December 2012.
- 1 2 Jacob, F.; Perrin, D.; Sánchez, C.; Monod, J. (Feb 1960). "L'opéron : groupe de gènes à expression coordonnée par un opérateur" [Operon: a group of genes with the expression coordinated by an operator] (PDF). Comptes rendus hebdomadaires des seances de l'Academie des sciences (Facsimile version reprinted in 2005) 250 (6): 1727–1729. ISSN 0001-4036. PMID 14406329.
- ↑ Miller JH, Suzuki DT, Griffiths AJF, Lewontin RC, Wessler SR, Gelbart WM (2005). Introduction to genetic analysis (8th ed.). San Francisco: W.H. Freeman. p. 740. ISBN 0-7167-4939-4.
- ↑ Jacob, F. O. (2011). "The Birth of the Operon". Science 332 (6031): 767–767. doi:10.1126/science.1207943. PMID 21566161.
- ↑ Blumenthal, Thomas (2004). "Operons in Eukaryotes". Brief Funct Genomic Proteomic 3 (3): 199–211. doi:10.1093/bfgp/3.3.199. PMID 15642184.
- 1 2 Lewin, Benjamin (1990). Genes IV (4th ed.). Oxford [Oxfordshire]: Oxford University Press. pp. 243–58. ISBN 0-19-854267-4.
- ↑ Mayer, Gene. "BACTERIOLOGY – CHAPTER NINE GENETIC REGULATORY MECHANISMS". Microbiology and Immunology Online. University of South Carolina School of Medicine. Retrieved 30 December 2012.
- ↑ Cummings MS, Klug WS (2006). Concepts of genetics (8th ed.). Upper Saddle River, NJ: Pearson Education. pp. 394–402. ISBN 0-13-191833-8.
- ↑ Salgado, H.; Moreno-Hagelsieb, G.; Smith, T.; Collado-Vides, J. (2000). "Operons in Escherichia coli: Genomic analyses and predictions". Proceedings of the National Academy of Sciences 97 (12): 6652–6657. doi:10.1073/pnas.110147297. PMC 18690. PMID 10823905.
- ↑ Ermolaeva, M.; White, O.; Salzberg, S. (2001). "Prediction of operons in microbial genomes". Nucleic Acids Research 29 (5): 1216–1221. doi:10.1093/nar/29.5.1216. PMC 29727. PMID 11222772.
- ↑ Toledo-Arana, A.; Dussurget, O.; Nikitas, G.; Sesto, N.; Guet-Revillet, H.; Balestrino, D.; Loh, E.; Gripenland, J.; Tiensuu, T.; Vaitkevicius, K.; Barthelemy, M.; Vergassola, M.; Nahori, M. A.; Soubigou, G.; Régnault, B. A.; Coppée, J. Y.; Lecuit, M.; Johansson, J. R.; Cossart, P. (2009). "The Listeria transcriptional landscape from saprophytism to virulence". Nature 459 (7249): 950–956. doi:10.1038/nature08080. PMID 19448609.
External links
- Mycobacterium tuberculosis H37Rv Operon Correlation Browser
- The Operon Concept: F.Jacob & J.Monad-1961
- OBD - Operon Database (a bit awkward to use though)
- Lac-operon
| ||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||