Multi-label classification
In machine learning, multi-label classification and the strongly related problem of multi-output classification are variants of the classification problem where multiple target labels must be assigned to each instance. Multi-label classification should not be confused with multiclass classification, which is the problem of categorizing instances into one of more than two classes. Formally, multi-label learning can be phrased as the problem of finding a model that maps inputs x to binary vectors y, rather than scalar outputs as in the ordinary classification problem.
There are two main methods for tackling the multi-label classification problem:[1] problem transformation methods and algorithm adaptation methods. Problem transformation methods transform the multi-label problem into a set of binary classification problems, which can then be handled using single-class classifiers. Algorithm adaptation methods adapt the algorithms to directly perform multi-label classification. In other words, rather than trying to convert the problem to a simpler problem, they try to address the problem in its full form.
Problem transformation methods
Several problem transformation methods exist for multi-label classification; the baseline approach, called the binary relevance method,[2][1][3] amounts to independently training one binary classifier for each label. Given an unseen sample, the combined model then predicts all labels for this sample for which the respective classifiers predict a positive result. This method of dividing the task into multiple binary tasks has something in common with the one-vs.-all (OvA, or one-vs.-rest, OvR) method for multiclass classification. Note though that it is not the same method: in binary relevance we train one classifier for each label, not one classifier for each possible value for the label.
Various other transformations exist. Of these, the label powerset (LP) transformation creates one binary classifier for every label combination attested in the training set.[1] The random k-labelsets (RAKEL) algorithm uses multiple LP classifiers, each trained on a random subset of the actual labels; prediction using this ensemble method proceeds by a voting scheme.[4]
Classifier chains are an alternative ensembling methods [2][3][5] that have been applied, for instance, in HIV drug resistance prediction.[6]
Adapted algorithms for multi-label classification
Some classification algorithms/models have been adaptated to the multi-label task, without requiring problem transformations. Examples of these include:
- boosting: AdaBoost.MH and AdaBoost.MR are extended versions of AdaBoost for multi-label data.
- k-nearest neighbors: the ML-kNN algorithm extends the k-NN classifier to multi-label data.[7]
- decision trees: "Clare" is an adapted C4.5 algorithm for multi-label classification; the modification involves the entropy calculations.[8]
- kernel methods for vector output
- neural networks: BP-MLL is an adaptation of the popular back-propagation algorithm for multi-label learning.[9]
Statistics and evaluation metrics
The extent to which a dataset is multi-label can be captured in two statistics:[1]
- Label cardinality is the average number of labels per example in the set:
 ;
; - label density is the number of labels per sample divided by the total number of labels, averaged over the samples:
 where
where  .
.
Evaluation metrics for multi-label classification performance are inherently different from those used in multi-class (or binary) classification, due to the inherent differences of the classification problem. If T denotes the true set of labels for a given sample, and P the predicted set of labels, then the following metrics can be defined on that sample:
- Hamming loss: the fraction of the wrong labels to the total number of labels, i.e.
 , where
, where 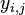 is the target and
is the target and 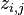 is the prediction. This is a loss function, so the optimal value is zero.
is the prediction. This is a loss function, so the optimal value is zero. - The closely related Hamming score, also called accuracy in the multi-label setting, is defined as the number of correct labels divided by the union of predicted and true labels,
 .[10]
.[10] - Precision, recall and
 score: precision is
score: precision is  , recall is
, recall is  , and
, and  is their harmonic mean.[10]
is their harmonic mean.[10] - Exact match: is the most strict metric, indicating the percentage of samples that have all their labels classified correctly.
Cross-validation in multi-label settings is complicated by the fact that the ordinary (binary/multiclass) way of stratified sampling will not work; alternative ways of approximate stratified sampling have been suggested.[11]
Implementations and datasets
Java implementations of multi-label algorithms are available in the Mulan and Meka software packages, both based on Weka.
The scikit-learn python package implements some multi-labels algorithms and metrics.
The binary relevance method with a lot of different base learners is implemented in the R-package mlr.
A list of commonly used multi-label data-sets is available at the Mulan website.
See also
References
- 1 2 3 4 Tsoumakas, Grigorios; Katakis, Ioannis (2007). "Multi-label classification: an overview" (PDF). International Journal of Data Warehousing & Mining 3 (3): 1–13. doi:10.4018/jdwm.2007070101.
- 1 2 Jesse Read, Bernhard Pfahringer, Geoff Holmes, Eibe Frank. Classifier Chains for Multi-label Classification. Machine Learning Journal. Springer. Vol. 85(3), (2011).
- 1 2 Read, Jesse; Martino, Luca; Luengo, David (2014-03-01). "Efficient monte carlo methods for multi-dimensional learning with classifier chains". Pattern Recognition. Handwriting Recognition and other PR Applications 47 (3): 1535–1546. doi:10.1016/j.patcog.2013.10.006.
- ↑ Tsoumakas, Grigorios; Vlahavas, Ioannis (2007). Random k-labelsets: An ensemble method for multilabel classification (PDF). ECML.
- ↑ Read, Jesse; Martino, Luca; Olmos, Pablo M.; Luengo, David (2015-06-01). "Scalable multi-output label prediction: From classifier chains to classifier trellises". Pattern Recognition 48 (6): 2096–2109. doi:10.1016/j.patcog.2015.01.004.
- ↑ Heider, D; Senge, R; Cheng, W; Hüllermeier, E (2013). "Multilabel classification for exploiting cross-resistance information in HIV-1 drug resistance prediction". Bioinformatics (Oxford, England) 29 (16): 1946–52. doi:10.1093/bioinformatics/btt331. PMID 23793752.
- ↑ Zhang, M.L.; Zhou, Z.H. (2007). "ML-KNN: A lazy learning approach to multi-label learning". Pattern Recognition 40 (7): 2038–2048. doi:10.1016/j.patcog.2006.12.019.
- ↑ Madjarov, Gjorgji; Kocev, Dragi; Gjorgjevikj, Dejan; Džeroski, Sašo (2012). "An extensive experimental comparison of methods for multi-label learning". Pattern Recognition 45 (9): 3084–3104. doi:10.1016/j.patcog.2012.03.004.
- ↑ Zhang, M.L.; Zhou, Z.H. (2006). Multi-label neural networks with applications to functional genomics and text categorization (PDF). IEEE Transactions on Knowledge and Data Engineering. pp. 1338–1351.
- 1 2 Godbole, Shantanu; Sarawagi, Sunita (2004). Discriminative methods for multi-labeled classification (PDF). Advances in Knowledge Discovery and Data Mining. pp. 22–30.
- ↑ Sechidis, Konstantinos; Tsoumakas, Grigorios; Vlahavas, Ioannis (2011). On the stratification of multi-label data (PDF). ECML PKDD. pp. 145–158.
Further reading
- Madjarov, Gjorgji; Kocev, Dragi; Gjorgjevikj, Dejan; Džeroski, Sašo (2012). "An extensive experimental comparison of methods for multi-label learning". Pattern Recognition 45 (9): 3084–3104. doi:10.1016/j.patcog.2012.03.004.