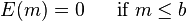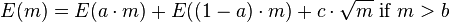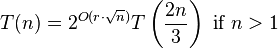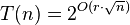Maximum disjoint set
In computational geometry, a maximum disjoint set (MDS) is a largest set of non-overlapping geometric shapes selected from a given set of candidate shapes.
Finding an MDS is important in applications such as automatic label placement, VLSI circuit design, and cellular frequency division multiplexing.
Every set of non-overlapping shapes is an independent set in the intersection graph of the shapes. Therefore, the MDS problem is a special case of the maximum independent set (MIS) problem. Both problems are NP complete, but finding a MDS may be easier than finding a MIS in two respects:
- For the general MIS problem, the best known exact algorithms are exponential. In some geometric intersection graphs, there are sub-exponential algorithms for finding a MDS.[1]
- The general MIS problem is hard to approximate and doesn't even have a constant-factor approximation. In some geometric intersection graphs, there are polynomial-time approximation schemes (PTAS) for finding a MDS.
The MDS problem can be generalized by assigning a different weight to each shape and searching for a disjoint set with a maximum total weight.
In the following text, MDS(C) denotes the maximum disjoint set in a set C.
Greedy algorithms
Given a set C of shapes, an approximation to MDS(C) can be found by the following greedy algorithm:
- INITIALIZATION: Initialize an empty set, S.
- SEARCH: For every shape x in C:
- Calculate N(x) - the subset of all shapes in C that intersect x (including x itself).
- Calculate the largest independent set in this subset: MDS(N(x)).
- Select an x such that |MDS(N(x))| is minimized.
- Add x to S.
- Remove x and N(x) from C.
- If there are shapes in C, go back to Search.
- END: return the set S.
For every shape x that we add to S, we lose the shapes in N(x), because they are intersected by x and thus cannot be added to S later on. However, some of these shapes themselves intersect each other, and thus in any case it is not possible that they all be in the optimal solution MDS(S). The largest subset of shapes that can all be in the optimal solution is MDS(N(x)). Therefore, selecting an x that minimizes |MDS(N(x))| minimizes the loss from adding x to S.
In particular, if we can guarantee that there is an x for which |MDS(N(x))| is bounded by a constant (say, M), then this greedy algorithm yields a constant M-factor approximation, as we can guarantee that:

Such an upper bound M exists for several interesting cases:
1-dimensional intervals: exact polynomial algorithm
When C is a set of intervals on a line, M=1, and thus the greedy algorithm finds the exact MDS. To see this, assume w.l.o.g. that the intervals are vertical, and let x be the interval with the highest bottom endpoint. All other intervals intersected by x must cross its bottom endpoint. Therefore, all intervals in N(x) intersect each other, and MDS(N(x)) has a size of at most 1 (see figure).
Therefore, in the 1-dimensional case, the MDS can be found exactly in time O(n log n):[2]
- Sort the intervals in ascending order of their bottom endpoints (this takes time O(n log n)).
- Add an interval with the highest bottom endpoint, and delete all intervals intersecting it.
- Continue until no intervals remain.
This algorithm is analogous to the earliest deadline first scheduling solution to the interval scheduling problem.
In contrast to the 1-dimensional case, in 2 or more dimensions the MDS problem becomes NP-complete, and thus has either exact super-polynomial algorithms or approximate polynomial algorithms.
Fat shapes: constant-factor approximations
When C is a set of unit disks, M=3,[3] because the leftmost disk (the disk whose center has the smallest x coordinate) intersects at most 3 other disjoint disks (see figure). Therefore the greedy algorithm yields a 3-approximation, i.e., it finds a disjoint set with a size of at least MDS(C)/3.
Similarly, when C is a set of axis-parallel unit squares, M=2.

When C is a set of arbitrary-size disks, M=5, because the disk with the smallest radius intersects at most 5 other disjoint disks (see figure).
Similarly, when C is a set of arbitrary-size axis-parallel squares, M=4.
Other constants can be calculated for other regular polygons.[3]
Divide-and-conquer algorithms
The most common approach to finding a MDS is divide-and-conquer. A typical algorithm in this approach looks like the following:
- Divide the given set of shapes into two or more subsets, such that the shapes in each subset cannot overlap the shapes in other subsets because of geometric considerations.
- Recursively find the MDS in each subset separately.
- Return the union of the MDSs from all subsets.
The main challenge with this approach is to find a geometric way to divide the set into subsets. This may require to discard a small number of shapes that do not fit into any one of the subsets, as explained in the following subsections.
Axis-parallel rectangles: Logarithmic-factor approximation
Let C be a set of n axis-parallel rectangles in the plane. The following algorithm finds a disjoint set with a size of at least 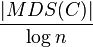 in time
in time
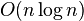 :[2]
:[2]
- INITIALIZATION: sort the horizontal edges of the given rectangles by their y-coordinate, and the vertical edges by their x-coordinate (this step takes time O(n log n)).
- STOP CONDITION: If there are at most n ≤ 2 shapes, compute the MDS directly and return.
- RECURSIVE PART:
- Let
 be the median x-coordinate.
be the median x-coordinate. - Partition the input rectangles into three groups according to their relation to the line
 : those entirely to its left (
: those entirely to its left ( ), those entirely to its right (
), those entirely to its right (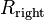 ), and those intersected by it (
), and those intersected by it (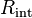 ). By construction, the cardinalities of
). By construction, the cardinalities of  and
and 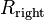 are at most n/2.
are at most n/2. - Recursively compute an approximate MDS in
 (
(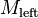 ) and in
) and in 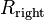 (
(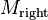 ), and calculate their union. By construction, the rectangles in
), and calculate their union. By construction, the rectangles in 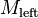 and
and 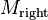 are all disjoint, so
are all disjoint, so 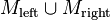 is a disjoint set.
is a disjoint set. - Compute an exact MDS in
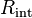 (
( ). Since all rectangles in
). Since all rectangles in 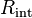 intersect a single vertical line
intersect a single vertical line  , this computation is equivalent to finding an MDS from a set of intervals, and can be solved exactly in time O(n log n) (see above).
, this computation is equivalent to finding an MDS from a set of intervals, and can be solved exactly in time O(n log n) (see above).
- Let
- Return either
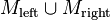 or
or  – whichever of them is larger.
– whichever of them is larger.
It is provable by induction that, at the last step, either 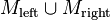 or
or  have a cardinality of at least
have a cardinality of at least 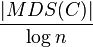 .
.
The approximation factor has recently been reduced to  [4] and generalized to the case in which rectangles have different weights.[5]
[4] and generalized to the case in which rectangles have different weights.[5]
Axis-parallel rectangles with the same height: 2-approximation
Let C be a set of n axis-parallel rectangles in the plane, all with the same height H but with varying lengths. The following algorithm finds a disjoint set with a size of at least |MDS(C)|/2 in time O(n log n):[2]
- Draw m horizontal lines, such that:
- The separation between two lines is strictly more than H.
- Each line intersects at least one rectangle (hence m ≤ n).
- Each rectangle is intersected by exactly one line.
- Since the height of all rectangles is H, it is not possible that a rectangle is intersected by more than one line. Therefore the lines partition the set of rectangles into m subsets (
 ) – each subset includes the rectangles intersected by a single line.
) – each subset includes the rectangles intersected by a single line. - For each subset
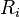 , compute an exact MDS
, compute an exact MDS  using the one-dimensional greedy algorithm (see above).
using the one-dimensional greedy algorithm (see above). - By construction, the rectangles in (
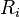 ) can intersect only rectangles in
) can intersect only rectangles in  or in
or in 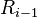 . Therefore, each of the following two unions is a disjoint sets:
. Therefore, each of the following two unions is a disjoint sets:
- Union of odd MDSs:
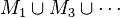
- Union of even MDSs:
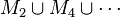
- Union of odd MDSs:
- Return the largest of these two unions. Its size must be at least |MDS|/2.
Axis-parallel rectangles with the same height: PTAS
Let C be a set of n axis-parallel rectangles in the plane, all with the same height but with varying lengths. There is an algorithm that finds a disjoint set with a size of at least |MDS(C)|/(1 + 1/k) in time O(n2k−1), for every constant k > 1.[2]
The algorithm is an improvement of the above-mentioned 2-approximation, by combining dynamic programming with the shifting technique of.[6]
This algorithm can be generalized to d dimensions. If the labels have the same size in all dimensions except one, it is possible to find a similar approximation by applying dynamic programming along one of the dimensions. This also reduces the time to n^O(1/e).[7]
Fat objects with identical sizes: PTAS
Let C be a set of n squares or circles of identical size. There is a polynomial-time approximation scheme for finding an MDS using a simple shifted-grid strategy. It finds a solution within (1 − e) of the maximum in time nO(1/e2) time and linear space.[6] The strategy generalizes to any collection of fat objects of roughly the same size (i.e., when the maximum-to-minimum size ratio is bounded by a constant).
Fat objects with arbitrary sizes: PTAS
Let C be a set of n fat objects (e.g. squares or circles) of arbitrary sizes. There is a PTAS for finding an MDS based on multi-level grid alignment. It has been discovered by two groups in approximately the same time, and described in two different ways.
Version 1 finds a disjoint set with a size of at least (1 − 1/k)2 · |MDS(C)| in time nO(k2), for every constant k > 1:[8]
Scale the disks so that the smallest disk has diameter 1. Partition the disks to levels, based on the logarithm of their size. I.e., the j-th level contains all disks with diameter between (k + 1)j and (k + 1)j+1, for j ≤ 0 (the smallest disk is in level 0).
For each level j, impose a grid on the plane that consists of lines that are (k + 1)j+1 apart from each other. By construction, every disk can intersect at most one horizontal line and one vertical line from its level.
For every r, s between 0 and k, define D(r,s) as the subset of disks that are not intersected by any horizontal line whose index modulo k is r, nor by any vertical line whose index modulu k is s. By the pigeonhole principle, there is at least one pair (r,s) such that  , i.e., we can find the MDS only in D(r,s) and miss only a small fraction of the disks in the optimal solution:
, i.e., we can find the MDS only in D(r,s) and miss only a small fraction of the disks in the optimal solution:
- For all k2 possible values of r,s (0 ≤ r,s < k), calculate D(r,s) using dynamic programming.
- Return the largest of these k2 sets.

Version 2 finds a disjoint set with a size of at least (1 − 2/k)·|MDS(C)| in time nO(k), for every constant k > 1.[7]
The algorithm uses shifted quadtrees. The key concept of the algorithm is alignment to the quadtree grid. An object of size r is called k-aligned (where k ≥ 1 is a constant) if it is inside a quadtree cell of size at most kr (R ≤ kr).
By definition, a k-aligned object that intersects the boundary of a quatree cell of size R must have a size of at least R/k (r > R/k). The boundary of a cell of size R can be covered by 4k squares of size R/k; hence the number of disjoint fat objects intersecting the boundary of that cell is at most 4kc, where c is a constant measuring the fatness of the objects.
Therefore, if all objects are fat and k-aligned, it is possible to find the exact maximum disjoint set in time nO(kc) using a divide-and-conquer algorithm. Start with a quadtree cell that contains all objects. Then recursively divide it to smaller quadtree cells, find the maximum in each smaller cell, and combine the results to get the maximum in the larger cell. Since the number of disjoint fat objects intersecting the boundary of every quadtree cell is bounded by 4kc, we can simply "guess" which objects intersect the boundary in the optimal solution, and then apply divide-and-conquer to the objects inside.
If almost all objects are k-aligned, we can just discard the objects that are not k-aligned, and find a maximum disjoint set of the remaining objects in time nO(k). This results in a (1 − e) approximation, where e is the fraction of objects that are not k-aligned.
If most objects are not k-aligned, we can try to make them k-aligned by shifting the grid in multiples of (1/k,1/k). First, scale the objects such that they are all contained in the unit square. Then, consider k shifts of the grid: (0,0), (1/k,1/k), (2/k,2/k), ..., ((k − 1)/k,(k − 1)/k). I.e., for each j in {0,...,k − 1}, consider a shift of the grid in (j/k,j/k). It is possible to prove that every label will be 2k-aligned for at least k − 2 values of j. Now, for every j, discard the objects that are not k-aligned in the (j/k,j/k) shift, and find a maximum disjoint set of the remaining objects. Call that set A(j). Call the real maximum disjoint set is A*. Then:

Therefore, the largest A(j) has a size of at least: (1 − 2/k)|A*|. The return value of the algorithm is the largest A(j); the approximation factor is (1 − 2/k), and the run time is nO(k). We can make the approximation factor as small as we want, so this is a PTAS.
Both versions can be generalized to d dimensions (with different approximation ratios) and to the weighted case.
Geometric separator algorithms
Several divide-and-conquer algorithms are based on a certain geometric separator theorem. A geometric separator is a line or shape that separates a given set of shapes to two smaller subsets, such that the number of shapes lost during the division is relatively small. This allows both PTASs and sub-exponential exact algorithms, as explained below.
Fat objects with arbitrary sizes: PTAS using geometric separators
Let C be a set of n fat objects of arbitrary sizes. The following algorithm finds a disjoint set with a size of at least (1 − O(√b))·|MDS(C)| in time nO(b), for every constant b > 1.[7]
The algorithm is based on the following geometric separator theorem, which can be proved similarly to the proof of the existence of geometric separator for disjoint squares:
- For every set C of fat objects, there is a rectangle that partitions C into three subsets of objects – Cinside, Coutside and Cboundary, such that:
- |MDS(Cinside)| ≤ a|MDS(C)|
- |MDS(Coutside)| ≤ a|MDS(C)|
- |MDS(Cboundary)| c√(|MDS(C)|)
- For every set C of fat objects, there is a rectangle that partitions C into three subsets of objects – Cinside, Coutside and Cboundary, such that:
where a and c are constants. If we could calculate MDS(C) exactly, we could make the constant a as low as 2/3 by a proper selection of the separator rectangle. But since we can only approximate MDS(C) by a constant factor, the constant a must be larger. Fortunately, a remains a constant independent of |C|.
This separator theorem allows to build the following PTAS:
Select a constant b. Check all possible combinations of up to b + 1 labels.
- If |MDS(C)| has a size of at most b (i.e. all sets of b + 1 labels are not disjoint) then just return that MDS and exit. This step takes nO(b) time.
- Otherwise, use a geometric separator to separate C to two subsets. Find the approximate MDS in Cinside and Coutside separately, and return their combination as the approximate MDS in C.
Let E(m) be the error of the above algorithm when the optimal MDS size is MDS(C) = m. When m ≤ b, the error is 0 because the maximum disjoint set is calculated exactly; when m > b, the error increases by at most c√m – the number of labels intersected by the separator. The worst case for the algorithm is when the split in each step is in the maximum possible ratio which is a:(1 − a). Therefore the error function satisfies the following recurrence relation:
The solution to this recurrence is:
i.e.,  . We can make the approximation factor as small as we want by a proper selection of b.
. We can make the approximation factor as small as we want by a proper selection of b.
This PTAS is more space-efficient than the PTAS based on quadtrees, and can handle a generalization where the objects may slide, but it cannot handle the weighted case.
Disks with a bounded size-ratio: exact sub-exponential algorithm
Let C be a set of n disks, such that the ratio between the largest radius and the smallest radius is at most r. The following algorithm finds MDS(C) exactly in time 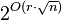 .[9]
.[9]
The algorithm is based on a width-bounded geometric separator on the set Q of the centers of all disks in C. This separator theorem allows to build the following exact algorithm:
- Find a separator line such that at most 2n/3 centers are to its right (Cright), at most 2n/3 centers are to its left (Cleft), and at most O(√n) centers are at a distance of less than r/2 from the line (Cint).
- Consider all possible non-overlapping subsets of Cint. There are at most
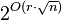 such subsets. For each such subset, recursively compute the MDS of Cleft and the MDS of Cright, and return the largest combined set.
such subsets. For each such subset, recursively compute the MDS of Cleft and the MDS of Cright, and return the largest combined set.
The run time of this algorithm satisfies the following recurrence relation:
The solution to this recurrence is:
Local search algorithms
Pseudo-disks: a PTAS
A pseudo-disks-set is a set of objects in which the boundaries of every pair of objects intersect at most twice. (Note that this definition relates to a whole collection, and does not say anything about the shapes of the specific objects in the collection). A pseudo-disks-set has a bounded union complexity, i.e., the number of intersection points on the boundary of the union of all objects is linear in the number of objects.
Let C be a pseudo-disks-set with n objects. The following local search algorithm finds a disjoint set of size at least 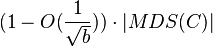 in time
in time 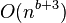 , for every integer constant
, for every integer constant  :[10]
:[10]
- INITIALIZATION: Initialize an empty set,
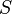 .
. - SEARCH: Loop over all the subsets of
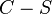 whose size is between 1 and
whose size is between 1 and 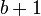 . For each such subset X:
. For each such subset X:
- Verify that X itself is independent (otherwise go to the next subset);
- Calculate the set Y of objects in S that intersect X.
- If
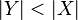 , then remove Y from S and insert X:
, then remove Y from S and insert X:  .
.
- END: return the set S.
Every exchange in the search step increases the size of S by at least 1, and thus can happen at most n times.
The algorithm is very simple; the difficult part is to prove the approximation ratio.[10]
See also.[11]
Linear programming relaxation algorithms
Pseudo-disks: a PTAS
Let C be a pseudo-disks-set with n objects and union complexity u. Using linear programming relaxation, it is possible to find a disjoint set of size at least 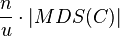 . This is possible either with a randomized algorithm that has a high probability of success and run time
. This is possible either with a randomized algorithm that has a high probability of success and run time  , or a deterministic algorithm with a slower run time (but still polynomial). This algorithm can be generalized to the weighted case.[10]
, or a deterministic algorithm with a slower run time (but still polynomial). This algorithm can be generalized to the weighted case.[10]
Other classes of shapes for which approximations are known
- Line segments and curves with a bounded number of intersection points.[12]
External links
- Approximation Algorithms for Maximum Independent Set of Pseudo-Disks - presentation by Sariel Har-Peled.
- Javascript code for calculating exact maximum disjoint set of rectangles.
Notes
- ↑ Ravi, S. S.; Hunt, H. B. (1987). "An application of the planar separator theorem to counting problems". Information Processing Letters 25 (5): 317. doi:10.1016/0020-0190(87)90206-7., Smith, W. D.; Wormald, N. C. (1998). "Geometric separator theorems and applications". Proceedings 39th Annual Symposium on Foundations of Computer Science (Cat. No.98CB36280). p. 232. doi:10.1109/sfcs.1998.743449. ISBN 0-8186-9172-7.
- 1 2 3 4 Agarwal, P. K.; Van Kreveld, M.; Suri, S. (1998). "Label placement by maximum independent set in rectangles". Computational Geometry 11 (3–4): 209. doi:10.1016/s0925-7721(98)00028-5.
- 1 2 Marathe, M. V.; Breu, H.; Hunt, H. B.; Ravi, S. S.; Rosenkrantz, D. J. (1995). "Simple heuristics for unit disk graphs". Networks 25 (2): 59. doi:10.1002/net.3230250205.
- ↑ Chalermsook, P.; Chuzhoy, J. (2009). "Maximum Independent Set of Rectangles". Proceedings of the Twentieth Annual ACM-SIAM Symposium on Discrete Algorithms. p. 892. doi:10.1137/1.9781611973068.97. ISBN 978-0-89871-680-1.
- ↑ Chalermsook, P. (2011). "Coloring and Maximum Independent Set of Rectangles". Approximation, Randomization, and Combinatorial Optimization. Algorithms and Techniques. Lecture Notes in Computer Science 6845. p. 123. doi:10.1007/978-3-642-22935-0_11. ISBN 978-3-642-22934-3.
- 1 2 Hochbaum, D. S.; Maass, W. (1985). "Approximation schemes for covering and packing problems in image processing and VLSI". Journal of the ACM 32: 130. doi:10.1145/2455.214106.
- 1 2 3 Chan, T. M. (2003). "Polynomial-time approximation schemes for packing and piercing fat objects". Journal of Algorithms 46 (2): 178–189. doi:10.1016/s0196-6774(02)00294-8.
- ↑ Erlebach, T.; Jansen, K.; Seidel, E. (2005). "Polynomial-Time Approximation Schemes for Geometric Intersection Graphs". SIAM Journal on Computing 34 (6): 1302. doi:10.1137/s0097539702402676.
- ↑ Fu, B. (2011). "Theory and application of width bounded geometric separators". Journal of Computer and System Sciences 77 (2): 379–392. doi:10.1016/j.jcss.2010.05.003.
- 1 2 3 Chan, T. M.; Har-Peled, S. (2012). "Approximation Algorithms for Maximum Independent Set of Pseudo-Disks". Discrete & Computational Geometry 48 (2): 373. doi:10.1007/s00454-012-9417-5.
- ↑ Agarwal, P. K.; Mustafa, N. H. (2006). "Independent set of intersection graphs of convex objects in 2D". Computational Geometry 34 (2): 83. doi:10.1016/j.comgeo.2005.12.001.
- ↑ Fox, J.; Pach, J. N. (2011). "Computing the Independence Number of Intersection Graphs". Proceedings of the Twenty-Second Annual ACM-SIAM Symposium on Discrete Algorithms. p. 1161. doi:10.1137/1.9781611973082.87. ISBN 978-0-89871-993-2.