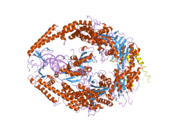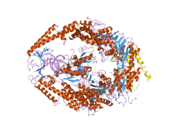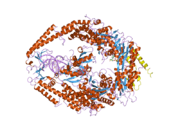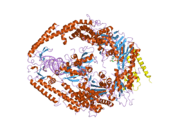MSH6
MSH6 or mutS homolog 6 is a gene that codes for DNA mismatch repair protein Msh6 in the budding yeast Saccharomyces cerevisiae. It is the homologue of the human "G/T binding protein," (GTBP) also called p160 or hMSH6 (human MSH6). The MSH6 protein is a member of the Mutator S (MutS) family of proteins that are involved in DNA damage repair.
Defects in hMSH6 are associated with atypical hereditary nonpolyposis colorectal cancer not fulfilling the Amsterdam criteria for HNPCC. hMSH6 mutations have also been linked to endometrial cancer and the development of endometrial carcinomas.
Discovery
MSH6 was first identified in the budding yeast S. cerevisiae because of its homology to MSH2. The identification of the human GTBP gene and subsequent amino acid sequence availability showed that yeast MSH6 and human GTBP were more related to each other than any other MutS homolog, with a 26.6% amino acid identity.[1] Thus, GTBP took on the name human MSH6, or hMSH6.
Structure
In the human genome, hMSH6 is located on chromosome 2. It contains the Walker-A/B adenine nucleotide binding motif, which is the most highly conserved sequence found in all MutS homologs.[2] As with other MutS homologs, hMSH6 has an intrinsic ATPase activity. It functions exclusively when bound to hMSH2 as a heterodimer, although hMSH2 itself can function as a homomultimer or as a heterodimer with hMSH3.[3]
Function
Importance of mismatch repair
Mismatches commonly occur as a result of DNA replication errors, genetic recombination, or other chemical and physical factors.[4] Recognizing those mismatches and repairing them is extremely important for cells, because failure to do so results in microsatellite instability, an elevated spontaneous mutation rate (mutator phenotype), and susceptibility to HNPCC.[2][5] hMSH6 combines with hMSH2 to form the active protein complex, hMutS alpha, also called hMSH2-hMSH6.
Mismatch recognition
Mismatch recognition by this complex is regulated by the ADP to ATP transformation, which provides evidence that hMutS alpha complex functions as a molecular switch.[6] In normal DNA, adenine (A) bonds with thymine (T) and cytosine (C) bonds with guanine (G). Sometimes there will be a mismatch where T will bind with G, which is called a G/T mismatch. When a G/T mismatch is recognized, hMutS alpha complex binds and exchanges ADP for ATP.[5] The ADP-->ATP exchange causes a conformational change to convert hMutS alpha into a sliding clamp that can diffuse along the DNA backbone.[5] The ATP induces a release of the complex from the DNA and allows the hMutS alpha to dissociate along the DNA like a sliding clamp. This transformation helps trigger downstream events to repair the damaged DNA.[5]
Cancer
Although mutations in hMSH2 cause a strong general mutator phenotype, mutations in hMSH6 cause only a modest mutator phenotype.[1] At the gene level, the mutations were found to cause primarily single-base substitution mutations, which suggests that the role of hMSH6 is primarily for correcting single-base substitution mutations and to a lesser extent single base insertion/deletion mutations.[1]
Mutations in the hMSH6 gene cause the protein to be nonfunctional or only partially active, thus reducing its ability to repair mistakes in DNA. The loss of MSH6 function results in instability at mononucleotide repeats.[1] HNPCC is most commonly caused by mutations in hMSH2 and hMLH1, but mutations in hMSH6 are linked to an atypical form of HNPCC.[7] The penetrance of colorectal cancer seems to be lower in these mutations, meaning that a low proportion of hMSH6 mutation carriers present with the disease. Endometrial cancer, on the other hand, seems to be a more important clinical manifestation for female mutation carriers. The onset of endometrial cancer and also colon cancer in families with hMSH6 mutations is about 50 years. This is delayed compared to the age 44 onset of hMSH2-related tumors.[7]
Interactions
MSH6 has been shown to interact with MSH2,[8][9][10][11][12] PCNA[13][14][15] and BRCA1.[8][16]
See also
References
- 1 2 3 4 Marsischky GT, et al. (1996) Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. ‘’Genes Dev.’’ 10(4):407-20. doi: 10.1101/gad.10.4.407. PMID 8600025
- 1 2 Fishel R, Kolodner RD. (1995). Identification of mismatch repair genes and their role in the development of cancer. Current Opinion in Genetics & Development. 5:382-95. doi: 10.1016/0959-437X(95)80055-7. PMID 7549435.
- ↑ Acharya S, et al. (1996) hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. ‘’PNAS.’’ 93(24):13629-34. doi: 10.1073/pnas.93.24.13629. PMID 8942985.
- ↑ Friedberg EC, Walker GC, Siede W. (1995). DNA repair and mutagenesis. American Society for Microbiology, Washington DC.
- 1 2 3 4 Gradia S, et al. (1999). hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. ‘’Molecular Cell.’’ 3(2):255-61. doi=10.1016/S1097-2765(00)80316-0. PMID 10078208
- ↑ Gradia S, Acharya S, Fishel R. (1997) The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. ‘’Cell.’’ 91:995-1005. doi: 10.1016/S0092-8674(00)80490-0. PMID 9428522.
- 1 2 Wagner, A; et al. (2001). Atypical HNPCC owing to MSH6 germline mutations: analysis of a large Dutch pedigree. ‘’J. Med. Genet.’’ 38(5):318–22. doi: 10.1136/jmg.38.5.318. PMID 11333868.
- 1 2 Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (Apr 2000). "BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures". Genes & Development 14 (8): 927–39. doi:10.1101/gad.14.8.927. PMC 316544. PMID 10783165.
- ↑ Wang Y, Qin J (Dec 2003). "MSH2 and ATR form a signaling module and regulate two branches of the damage response to DNA methylation". Proceedings of the National Academy of Sciences of the United States of America 100 (26): 15387–92. doi:10.1073/pnas.2536810100. PMC 307577. PMID 14657349.
- ↑ Guerrette S, Wilson T, Gradia S, Fishel R (Nov 1998). "Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6: examination of mutations found in hereditary nonpolyposis colorectal cancer". Molecular and Cellular Biology 18 (11): 6616–23. doi:10.1128/mcb.18.11.6616. PMC 109246. PMID 9774676.
- ↑ Bocker T, Barusevicius A, Snowden T, Rasio D, Guerrette S, Robbins D, Schmidt C, Burczak J, Croce CM, Copeland T, Kovatich AJ, Fishel R (Feb 1999). "hMSH5: a human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis". Cancer Research 59 (4): 816–22. PMID 10029069.
- ↑ Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R (Nov 1996). "hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6". Proceedings of the National Academy of Sciences of the United States of America 93 (24): 13629–34. doi:10.1073/pnas.93.24.13629. PMC 19374. PMID 8942985.
- ↑ Kleczkowska HE, Marra G, Lettieri T, Jiricny J (Mar 2001). "hMSH3 and hMSH6 interact with PCNA and colocalize with it to replication foci". Genes & Development 15 (6): 724–36. doi:10.1101/gad.191201. PMC 312660. PMID 11274057.
- ↑ Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA (Nov 2000). "Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes". The Journal of Biological Chemistry 275 (47): 36498–501. doi:10.1074/jbc.C000513200. PMID 11005803.
- ↑ Ohta S, Shiomi Y, Sugimoto K, Obuse C, Tsurimoto T (Oct 2002). "A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2-5 complex as a novel PCNA-binding protein". The Journal of Biological Chemistry 277 (43): 40362–7. doi:10.1074/jbc.M206194200. PMID 12171929.
- ↑ Wang Q, Zhang H, Guerrette S, Chen J, Mazurek A, Wilson T, Slupianek A, Skorski T, Fishel R, Greene MI (Aug 2001). "Adenosine nucleotide modulates the physical interaction between hMSH2 and BRCA1". Oncogene 20 (34): 4640–9. doi:10.1038/sj.onc.1204625. PMID 11498787.
Further reading
- Drummond JT, Li GM, Longley MJ, Modrich P (Jun 1995). "Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells". Science 268 (5219): 1909–12. doi:10.1126/science.7604264. PMID 7604264.
- Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D'Arrigo A, Truong O, Hsuan JJ, Jiricny J (Jun 1995). "GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells". Science 268 (5219): 1912–4. doi:10.1126/science.7604265. PMID 7604265.
- Papadopoulos N, Nicolaides NC, Liu B, Parsons R, Lengauer C, Palombo F, D'Arrigo A, Markowitz S, Willson JK, Kinzler KW (Jun 1995). "Mutations of GTBP in genetically unstable cells". Science 268 (5219): 1915–7. doi:10.1126/science.7604266. PMID 7604266.
- Risinger JI, Umar A, Boyd J, Berchuck A, Kunkel TA, Barrett JC (Sep 1996). "Mutation of MSH3 in endometrial cancer and evidence for its functional role in heteroduplex repair". Nature Genetics 14 (1): 102–5. doi:10.1038/ng0996-102. PMID 8782829.
- Nicolaides NC, Palombo F, Kinzler KW, Vogelstein B, Jiricny J (Feb 1996). "Molecular cloning of the N-terminus of GTBP". Genomics 31 (3): 395–7. doi:10.1006/geno.1996.0067. PMID 8838326.
- Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, Kolodner R, Fishel R (Nov 1996). "hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6". Proceedings of the National Academy of Sciences of the United States of America 93 (24): 13629–34. doi:10.1073/pnas.93.24.13629. PMC 19374. PMID 8942985.
- Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, Igari T, Koike M, Chiba M, Mori T (Nov 1997). "Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer". Nature Genetics 17 (3): 271–2. doi:10.1038/ng1197-271. PMID 9354786.
- Yin J, Kong D, Wang S, Zou TT, Souza RF, Smolinski KN, Lynch PM, Hamilton SR, Sugimura H, Powell SM, Young J, Abraham JM, Meltzer SJ (1998). "Mutation of hMSH3 and hMSH6 mismatch repair genes in genetically unstable human colorectal and gastric carcinomas". Human Mutation 10 (6): 474–8. doi:10.1002/(SICI)1098-1004(1997)10:6<474::AID-HUMU9>3.0.CO;2-D. PMID 9401011.
- Gradia S, Acharya S, Fishel R (Dec 1997). "The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch". Cell 91 (7): 995–1005. doi:10.1016/S0092-8674(00)80490-0. PMID 9428522.
- Shiwaku HO, Wakatsuki S, Mori Y, Fukushige S, Horii A (Oct 1997). "Alternative splicing of GTBP in normal human tissues". DNA Research 4 (5): 359–62. doi:10.1093/dnares/4.5.359. PMID 9455487.
- Wei Q, Guan Y, Cheng L, Radinsky R, Bar-Eli M, Tsan R, Li L, Legerski RJ (1998). "Expression of five selected human mismatch repair genes simultaneously detected in normal and cancer cell lines by a nonradioactive multiplex reverse transcription-polymerase chain reaction". Pathobiology 65 (6): 293–300. doi:10.1159/000164141. PMID 9491849.
- Guerrette S, Wilson T, Gradia S, Fishel R (Nov 1998). "Interactions of human hMSH2 with hMSH3 and hMSH2 with hMSH6: examination of mutations found in hereditary nonpolyposis colorectal cancer". Molecular and Cellular Biology 18 (11): 6616–23. doi:10.1128/mcb.18.11.6616. PMC 109246. PMID 9774676.
- Wang Q, Lasset C, Desseigne F, Saurin JC, Maugard C, Navarro C, Ruano E, Descos L, Trillet-Lenoir V, Bosset JF, Puisieux A (1999). "Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer". Human Genetics 105 (1-2): 79–85. doi:10.1007/s004390051067. PMID 10480359.
- Wijnen J, de Leeuw W, Vasen H, van der Klift H, Møller P, Stormorken A, Meijers-Heijboer H, Lindhout D, Menko F, Vossen S, Möslein G, Tops C, Bröcker-Vriends A, Wu Y, Hofstra R, Sijmons R, Cornelisse C, Morreau H, Fodde R (Oct 1999). "Familial endometrial cancer in female carriers of MSH6 germline mutations". Nature Genetics 23 (2): 142–4. doi:10.1038/13773. PMID 10508506.
- Wu Y, Berends MJ, Mensink RG, Kempinga C, Sijmons RH, van Der Zee AG, Hollema H, Kleibeuker JH, Buys CH, Hofstra RM (Nov 1999). "Association of hereditary nonpolyposis colorectal cancer-related tumors displaying low microsatellite instability with MSH6 germline mutations". American Journal of Human Genetics 65 (5): 1291–8. doi:10.1086/302612. PMC 1288281. PMID 10521294.
- Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, Wahlberg S, Fox EA, Peel D, Ziogas A, Garber JE, Syngal S, Anton-Culver H, Li FP (Oct 1999). "Germ-line msh6 mutations in colorectal cancer families". Cancer Research 59 (20): 5068–74. PMID 10537275.
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (Apr 2000). "BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures". Genes & Development 14 (8): 927–39. doi:10.1101/gad.14.8.927. PMC 316544. PMID 10783165.
- Ceccotti S, Ciotta C, Fronza G, Dogliotti E, Bignami M (Jul 2000). "Multiple mutations and frameshifts are the hallmark of defective hPMS2 in pZ189-transfected human tumor cells". Nucleic Acids Research 28 (13): 2577–84. doi:10.1093/nar/28.13.2577. PMC 102707. PMID 10871409.
- Christmann M, Kaina B (Nov 2000). "Nuclear translocation of mismatch repair proteins MSH2 and MSH6 as a response of cells to alkylating agents". The Journal of Biological Chemistry 275 (46): 36256–62. doi:10.1074/jbc.M005377200. PMID 10954713.
- Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA (Nov 2000). "Functional interaction of proliferating cell nuclear antigen with MSH2-MSH6 and MSH2-MSH3 complexes". The Journal of Biological Chemistry 275 (47): 36498–501. doi:10.1074/jbc.C000513200. PMID 11005803.
External links
- FAQs on HNPCC from the National Institute of Health
- GeneReviews/NCBI/NIH/UW entry on Lynch syndrome
- MSH6 protein, human at the US National Library of Medicine Medical Subject Headings (MeSH)
| |||||||||||||||||||