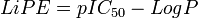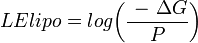Lipophilic efficiency
Lipophilic efficiency[1] (LiPE), sometimes referred to as ligand-lipophilicity efficiency (LLE) is a parameter used in drug design and drug discovery to evaluate the quality of research compounds, linking potency and lipophilicity in an attempt to estimate druglikeness.[2][3] For a given compound LiPE is defined as the pIC50 (or pEC50) of interest minus the LogP of the compound.
In practice, the calculated value cLogP is often used instead of the measured LogP. LiPE is used to compare compounds of different potencies (pIC50s) and lipophilicities (LogP). High potency (high value of pIC50) is a desirable attribute in drug candidates, as it reduces the risk of non-specific, off-target pharmacology at a given concentration. When associated with low clearance, high potency also allows for low total dose, which lowers the risk of idiosyncratic drug reaction.[4][5]
On the other hand, LogP is an estimate of a compound's overall lipophilicity, a value that influence its behavior in a range of biological processes relevant to a drug discovery, such as solubility, permeability through biological membranes, hepatic clearance, lack of selectivity and non-specific toxicity.[6] For oral drugs, a LogP value comprised between 2 and 3 is often considered optimal to achieve a compromise between permeability and first-pass clearance.
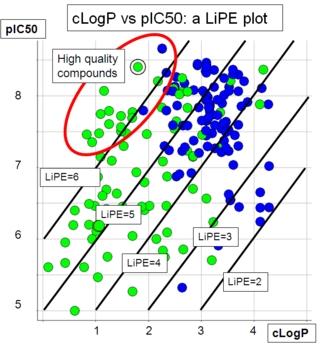
LiPE allows to capture both values in a single parameter, and empirical evidence suggest that quality drug candidates have a high LiPE (>6); this value corresponds to a compound with a pIC50 of 8 and a LogP of 2. Plotting LogP against pIC50 for a range of compounds allows to rank series and individual compounds.
An alternative equation uses the logarithm of the ratio of potency (measured as binding energy) and the partition coefficient to compute a lipophilic ligand efficiency index (LE) with a different scale.[7]
The following review discusses LipE in the context of other compound efficiency metrics.[8]
References
- ↑ Ryckmans T, Edwards MP, Horne VA, Correia AM, Owen DR, Thompson LR, Tran I, Tutt MF, Young T (August 2009). "Rapid assessment of a novel series of selective CB(2) agonists using parallel synthesis protocols: A Lipophilic Efficiency (LipE) analysis". Bioorg. Med. Chem. Lett. 19 (15): 4406–9. doi:10.1016/j.bmcl.2009.05.062. PMID 19500981.
- ↑ Edwards MP, Price DA (2010). "Role of Physicochemical Properties and Ligand Lipophilicity Efficiency in Addressing Drug Safety Risks". Annual Reports in Medicinal Chemistry 45: 381–391. doi:10.1016/S0065-7743(10)45023-X.
- ↑ Leeson PD, Springthorpe B (November 2007). "The influence of drug-like concepts on decision-making in medicinal chemistry". Nat Rev Drug Discov 6 (11): 881–90. doi:10.1038/nrd2445. PMID 17971784.
- ↑ Uetrecht J (January 2001). "Prediction of a new drug's potential to cause idiosyncratic reactions". Curr Opin Drug Discov Devel 4 (1): 55–9. PMID 11727323.
- ↑ Uetrecht J (January 2008). "Idiosyncratic drug reactions: past, present, and future". Chem. Res. Toxicol. 21 (1): 84–92. doi:10.1021/tx700186p. PMID 18052104.
- ↑ Hughes JD, Blagg J, Price DA, Bailey S, Decrescenzo GA, Devraj RV, Ellsworth E, Fobian YM, Gibbs ME, Gilles RW, Greene N, Huang E, Krieger-Burke T, Loesel J, Wager T, Whiteley L, Zhang Y (September 2008). "Physiochemical drug properties associated with in vivo toxicological outcomes". Bioorg. Med. Chem. Lett. 18 (17): 4872–5. doi:10.1016/j.bmcl.2008.07.071. PMID 18691886.
- ↑ Garcia-Sosa AT, Hetenyi C, Maran U (2010). "Drug efficiency indices for improvement of molecular docking scoring functions". Journal of Computational Chemistry 31: 174–184. doi:10.1002/jcc.21306. PMID 19422000.
- ↑ Shultz MD (2013). "Setting expectations in molecular optimizations: Strengths and limitations of commonly used composite parameters". Bioorg. Med. Chem. Lett. 23 (21): 5980–91. doi:10.1016/j.bmcl.2013.08.029. PMID 24018190.
| ||||||