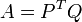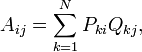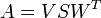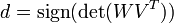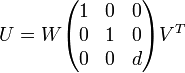Kabsch algorithm
The Kabsch algorithm, named after Wolfgang Kabsch, is a method for calculating the optimal rotation matrix that minimizes the RMSD (root mean squared deviation) between two paired sets of points. It is useful in graphics, cheminformatics to compare molecular structures, and also bioinformatics for comparing protein structures (in particular, see root-mean-square deviation (bioinformatics)).
The algorithm only computes the rotation matrix, but it also requires the computation of a translation vector. When both the translation and rotation are actually performed, the algorithm is sometimes called partial Procrustes superimposition (see also orthogonal Procrustes problem).
Description
The algorithm starts with two sets of paired points, P and Q. Each set of points can be represented as an N×3 matrix. The first row is the coordinates of the first point, the second row is the coordinates of the second point, the Nth row is the coordinates of the Nth point.
The algorithm works in three steps: a translation, the computation of a covariance matrix, and the computation of the optimal rotation matrix.
Translation
Both sets of coordinates must be translated first, so that their centroid coincides with the origin of the coordinate system. This is done by subtracting from the point coordinates the coordinates of the respective centroid.
Computation of the covariance matrix
The second step consist of calculating a covariance matrix A. In matrix notation,
or, using summation notation,
Computation of the optimal rotation matrix
It is possible to calculate the optimal rotation U based on the matrix formula 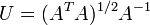 but implementing a numerical solution to this formula becomes complicated when all special cases are accounted for (for example, the case of A not having an inverse).
but implementing a numerical solution to this formula becomes complicated when all special cases are accounted for (for example, the case of A not having an inverse).
If singular value decomposition (SVD) routines are available, the optimal rotation, U, can be calculated using the following simple algorithm.
First, calculate the SVD of the covariance matrix A.
Next, decide whether we need to correct our rotation matrix to ensure a right-handed coordinate system
Finally, calculate our optimal rotation matrix, U, as
Coutsias, Seok, and Dill[1] have found an equivalent method that uses quaternions. Expressing the optimal rotation matrix with a quaternion goes back to 1999: see appendix in [2] and was generalized in 2002 to probability distributions (continuous or not): see appendix A.5 in.[3]
Generalizations
The algorithm was described for points in a three-dimensional space. The generalization to D dimensions is immediate.
External links
This SVD algorithm is described in more detail at http://cnx.org/content/m11608/latest/
A Matlab function is available at http://www.mathworks.com/matlabcentral/fileexchange/25746-kabsch-algorithm
A C++ implementation (and unit test) using Eigen
A Python script is available at https://github.com/charnley/rmsd
A free PyMol plugin easily implementing Kabsch is Cealign. VMD uses the Kabsch algorithm for its alignment.
See also
References
- ↑ Coutsias EA, Seok C, Dill KA (2004). "Using quaternions to calculate RMSD". J Comput Chem 25 (15): 1849–1857. doi:10.1002/jcc.20110. PMID 15376254.
- ↑ Petitjean M (1999). "On the root mean square quantitative chirality and quantitative symmetry measures". J Math Phys 40 (9): 4587–4595. doi:10.1063/1.532988.
- ↑ Petitjean M (2002). "Chiral mixtures". J Math Phys 43 (8): 4147–4157. doi:10.1063/1.1484559.
- Kabsch, Wolfgang, (1976) "A solution for the best rotation to relate two sets of vectors", Acta Crystallographica 32:922. doi:10.1107/S0567739476001873 with a correction in Kabsch, Wolfgang, (1978) "A discussion of the solution for the best rotation to relate two sets of vectors", "Acta Crystallographica", "A34", 827–828 doi:10.1107/S0567739478001680
- Lin Ying-Hung, Chang Hsun-Chang, Lin Yaw-Ling (2004) "A Study on Tools and Algorithms for 3-D Protein Structures Alignment and Comparison", International Computer Symposium, December 15–17, Taipei, Taiwan.
- Umeyama, Shinj, (1991) "Least-Squares Estimation of Transformation Parameters Between Two Point Patterns". IEEE Trans. Pattern Anal. Mach. Intell.. 13(4):376-380 doi:10.1109/34.88573