k-means clustering
| Machine learning and data mining |
|---|
 |
| Problems |
| Clustering |
| Dimensionality reduction |
| Structured prediction |
| Anomaly detection |
| Neural nets |
| Theory |
| Machine learning venues |
| Data mining venues |
|
|
k-means clustering is a method of vector quantization, originally from signal processing, that is popular for cluster analysis in data mining. k-means clustering aims to partition n observations into k clusters in which each observation belongs to the cluster with the nearest mean, serving as a prototype of the cluster. This results in a partitioning of the data space into Voronoi cells.
The problem is computationally difficult (NP-hard); however, there are efficient heuristic algorithms that are commonly employed and converge quickly to a local optimum. These are usually similar to the expectation-maximization algorithm for mixtures of Gaussian distributions via an iterative refinement approach employed by both algorithms. Additionally, they both use cluster centers to model the data; however, k-means clustering tends to find clusters of comparable spatial extent, while the expectation-maximization mechanism allows clusters to have different shapes.
The algorithm has a loose relationship to the k-nearest neighbor classifier, a popular machine learning technique for classification that is often confused with k-means because of the k in the name. One can apply the 1-nearest neighbor classifier on the cluster centers obtained by k-means to classify new data into the existing clusters. This is known as nearest centroid classifier or Rocchio algorithm.
Description
Given a set of observations (x1, x2, …, xn), where each observation is a d-dimensional real vector, k-means clustering aims to partition the n observations into k (≤ n) sets S = {S1, S2, …, Sk} so as to minimize the within-cluster sum of squares (WCSS) (sum of distance functions of each point in the cluster to the K center). In other words, its objective is to find:
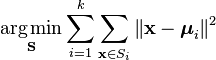
where μi is the mean of points in Si.
History
The term "k-means" was first used by James MacQueen in 1967,[1] though the idea goes back to Hugo Steinhaus in 1957.[2] The standard algorithm was first proposed by Stuart Lloyd in 1957 as a technique for pulse-code modulation, though it wasn't published outside of Bell Labs until 1982.[3] In 1965, E.W.Forgy published essentially the same method, which is why it is sometimes referred to as Lloyd-Forgy.[4] A more efficient version was proposed and published in Fortran by Hartigan and Wong in 1975/1979.[5][6]
Algorithms
Standard algorithm
The most common algorithm uses an iterative refinement technique. Due to its ubiquity it is often called the k-means algorithm; it is also referred to as Lloyd's algorithm, particularly in the computer science community.
Given an initial set of k means m1(1),…,mk(1) (see below), the algorithm proceeds by alternating between two steps:[7]
- Assignment step: Assign each observation to the cluster whose mean yields the least within-cluster sum of squares (WCSS). Since the sum of squares is the squared Euclidean distance, this is intuitively the "nearest" mean.[8] (Mathematically, this means partitioning the observations according to the Voronoi diagram generated by the means).

- where each
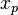 is assigned to exactly one
is assigned to exactly one  , even if it could be assigned to two or more of them.
, even if it could be assigned to two or more of them.
- Update step: Calculate the new means to be the centroids of the observations in the new clusters.
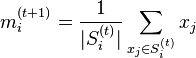
- Since the arithmetic mean is a least-squares estimator, this also minimizes the within-cluster sum of squares (WCSS) objective.
The algorithm has converged when the assignments no longer change. Since both steps optimize the WCSS objective, and there only exists a finite number of such partitionings, the algorithm must converge to a (local) optimum. There is no guarantee that the global optimum is found using this algorithm.
The algorithm is often presented as assigning objects to the nearest cluster by distance. The standard algorithm aims at minimizing the WCSS objective, and thus assigns by "least sum of squares", which is exactly equivalent to assigning by the smallest Euclidean distance. Using a different distance function other than (squared) Euclidean distance may stop the algorithm from converging. Various modifications of k-means such as spherical k-means and k-medoids have been proposed to allow using other distance measures.
Initialization methods
Commonly used initialization methods are Forgy and Random Partition.[9] The Forgy method randomly chooses k observations from the data set and uses these as the initial means. The Random Partition method first randomly assigns a cluster to each observation and then proceeds to the update step, thus computing the initial mean to be the centroid of the cluster's randomly assigned points. The Forgy method tends to spread the initial means out, while Random Partition places all of them close to the center of the data set. According to Hamerly et al.,[9] the Random Partition method is generally preferable for algorithms such as the k-harmonic means and fuzzy k-means. For expectation maximization and standard k-means algorithms, the Forgy method of initialization is preferable.
- Demonstration of the standard algorithm
-

1. k initial "means" (in this case k=3) are randomly generated within the data domain (shown in color).
-

2. k clusters are created by associating every observation with the nearest mean. The partitions here represent the Voronoi diagram generated by the means.
-

3. The centroid of each of the k clusters becomes the new mean.
-

4. Steps 2 and 3 are repeated until convergence has been reached.
As it is a heuristic algorithm, there is no guarantee that it will converge to the global optimum, and the result may depend on the initial clusters. As the algorithm is usually very fast, it is common to run it multiple times with different starting conditions. However, in the worst case, k-means can be very slow to converge: in particular it has been shown that there exist certain point sets, even in 2 dimensions, on which k-means takes exponential time, that is 2Ω(n), to converge.[10] These point sets do not seem to arise in practice: this is corroborated by the fact that the smoothed running time of k-means is polynomial.[11]
The "assignment" step is also referred to as expectation step, the "update step" as maximization step, making this algorithm a variant of the generalized expectation-maximization algorithm.
Complexity
Regarding computational complexity, finding the optimal solution to the k-means clustering problem for observations in d dimensions is:
- NP-hard in general Euclidean space d even for 2 clusters[12][13]
- NP-hard for a general number of clusters k even in the plane[14]
- If k and d (the dimension) are fixed, the problem can be exactly solved in time
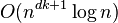 , where n is the number of entities to be clustered[15]
, where n is the number of entities to be clustered[15]
Thus, a variety of heuristic algorithms such as Lloyds algorithm given above are generally used.
The running time of Lloyds algorithm is often given as 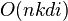 , where n is the number of d-dimensional vectors, k the number of clusters and i the number of iterations needed until convergence. On data that does have a clustering structure, the number of iterations until convergence is often small, and results only improve slightly after the first dozen iterations. Lloyds algorithm is therefore often considered to be of "linear" complexity in practice.
, where n is the number of d-dimensional vectors, k the number of clusters and i the number of iterations needed until convergence. On data that does have a clustering structure, the number of iterations until convergence is often small, and results only improve slightly after the first dozen iterations. Lloyds algorithm is therefore often considered to be of "linear" complexity in practice.
Following are some recent insights into this algorithm complexity behavior.
- Lloyd's k-means algorithm has polynomial smoothed running time. It is shown that[11] for arbitrary set of n points in
![[0,1]^d](../I/m/26bc47a1ae7c57f9c370dd5928d898a4.png) , if each point is independently perturbed by a normal distribution with mean 0 and variance
, if each point is independently perturbed by a normal distribution with mean 0 and variance  , then the expected running time of k-means algorithm is bounded by
, then the expected running time of k-means algorithm is bounded by  , which is a polynomial in n, k, d and
, which is a polynomial in n, k, d and  .
. - Better bounds are proved for simple cases. For example,[16] showed that the running time of k-means algorithm is bounded by
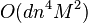 for n points in an integer lattice
for n points in an integer lattice 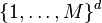 .
.
Lloyd's algorithm is the standard approach for this problem, However, it spends a lot of processing time computing the distances between each of the k cluster centers and the n data points. Since points usually stay in the same clusters after a few iterations, much of this work is unnecessary, making the naive implementation very inefficient. Some implementations use the triangle inequality in order to create bounds and accelerate Lloyds algorithm.[17][18][19]
Variations
- Jenks natural breaks optimization: k-means applied to univariate data
- k-medians clustering uses the median in each dimension instead of the mean, and this way minimizes
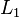 norm (Taxicab geometry).
norm (Taxicab geometry). - k-medoids (also: Partitioning Around Medoids, PAM) uses the medoid instead of the mean, and this way minimizes the sum of distances for arbitrary distance functions.
- Fuzzy C-Means Clustering is a soft version of K-means, where each data point has a fuzzy degree of belonging to each cluster.
- Gaussian mixture models trained with expectation-maximization algorithm (EM algorithm) maintains probabilistic assignments to clusters, instead of deterministic assignments, and multivariate Gaussian distributions instead of means.
- k-means++ chooses initial centers in a way that gives a provable upper bound on the WCSS objective.
- The filtering algorithm uses kd-trees to speed up each k-means step.[20]
- Some methods attempt to speed up each k-means step using the triangle inequality.[17][18][19][21]
- Escape local optima by swapping points between clusters.[6]
- The Spherical k-means clustering algorithm is suitable for directional data.[22]
- X-means clustering and G-means clustering try to automatically determine the number of clusters.
- Internal cluster evaluation measures such as cluster silhouette can be helpful at determining the number of clusters.
- Minkowski weighted k-means automatically calculates cluster specific feature weights, supporting the intuitive idea that a feature may have different degrees of relevance at different features.[23] These weights can also be used to re-scale a given data set, increasing the likelihood of a cluster validity index to be optimized at the expected number of clusters.[24]
Discussion



Three key features of k-means which make it efficient are often regarded as its biggest drawbacks:
- Euclidean distance is used as a metric and variance is used as a measure of cluster scatter.
- The number of clusters k is an input parameter: an inappropriate choice of k may yield poor results. That is why, when performing k-means, it is important to run diagnostic checks for determining the number of clusters in the data set.
- Convergence to a local minimum may produce counterintuitive ("wrong") results (see example in Fig.).
A key limitation of k-means is its cluster model. The concept is based on spherical clusters that are separable in a way so that the mean value converges towards the cluster center. The clusters are expected to be of similar size, so that the assignment to the nearest cluster center is the correct assignment. When for example applying k-means with a value of  onto the well-known Iris flower data set, the result often fails to separate the three Iris species contained in the data set. With
onto the well-known Iris flower data set, the result often fails to separate the three Iris species contained in the data set. With  , the two visible clusters (one containing two species) will be discovered, whereas with
, the two visible clusters (one containing two species) will be discovered, whereas with  one of the two clusters will be split into two even parts. In fact,
one of the two clusters will be split into two even parts. In fact,  is more appropriate for this data set, despite the data set containing 3 classes. As with any other clustering algorithm, the k-means result relies on the data set to satisfy the assumptions made by the clustering algorithms. It works well on some data sets, while failing on others.
is more appropriate for this data set, despite the data set containing 3 classes. As with any other clustering algorithm, the k-means result relies on the data set to satisfy the assumptions made by the clustering algorithms. It works well on some data sets, while failing on others.
The result of k-means can also be seen as the Voronoi cells of the cluster means. Since data is split halfway between cluster means, this can lead to suboptimal splits as can be seen in the "mouse" example. The Gaussian models used by the Expectation-maximization algorithm (which can be seen as a generalization of k-means) are more flexible here by having both variances and covariances. The EM result is thus able to accommodate clusters of variable size much better than k-means as well as correlated clusters (not in this example).
Applications
k-means clustering, in particular when using heuristics such as Lloyd's algorithm, is rather easy to implement and apply even on large data sets. As such, it has been successfully used in various topics, including market segmentation, computer vision, geostatistics,[26] astronomy and agriculture. It often is used as a preprocessing step for other algorithms, for example to find a starting configuration.
Vector quantization


k-means originates from signal processing, and still finds use in this domain. For example in computer graphics, color quantization is the task of reducing the color palette of an image to a fixed number of colors k. The k-means algorithm can easily be used for this task and produces competitive results. A use case for this approach is image segmentation. Other uses of vector quantization include non-random sampling, as k-means can easily be used to choose k different but prototypical objects from a large data set for further analysis.
Cluster analysis
In cluster analysis, the k-means algorithm can be used to partition the input data set into k partitions (clusters).
However, the pure k-means algorithm is not very flexible, and as such is of limited use (except for when vector quantization as above is actually the desired use case!). In particular, the parameter k is known to be hard to choose (as discussed above) when not given by external constraints. Another limitation of the algorithm is that it cannot be used with arbitrary distance functions or on non-numerical data. For these use cases, many other algorithms have been developed since.
Feature learning
k-means clustering has been used as a feature learning (or dictionary learning) step, in either (semi-)supervised learning or unsupervised learning.[27] The basic approach is first to train a k-means clustering representation, using the input training data (which need not be labelled). Then, to project any input datum into the new feature space, we have a choice of "encoding" functions, but we can use for example the thresholded matrix-product of the datum with the centroid locations, the distance from the datum to each centroid, or simply an indicator function for the nearest centroid,[27][28] or some smooth transformation of the distance.[29] Alternatively, by transforming the sample-cluster distance through a Gaussian RBF, one effectively obtains the hidden layer of a radial basis function network.[30]
This use of k-means has been successfully combined with simple, linear classifiers for semi-supervised learning in NLP (specifically for named entity recognition)[31] and in computer vision. On an object recognition task, it was found to exhibit comparable performance with more sophisticated feature learning approaches such as autoencoders and restricted Boltzmann machines.[29] However, it generally requires more data than the sophisticated methods, for equivalent performance, because each data point only contributes to one "feature" rather than multiple.[27]
Relation to other statistical machine learning algorithms
k-means clustering, and its associated expectation-maximization algorithm, is a special case of a Gaussian mixture model, specifically, the limit of taking all covariances as diagonal, equal, and small. It is often easy to generalize a k-means problem into a Gaussian mixture model.[32] Another generalization of the k-means algorithm is the K-SVD algorithm, which estimates data points as a sparse linear combination of "codebook vectors". K-means corresponds to the special case of using a single codebook vector, with a weight of 1.[33]
Mean shift clustering
Basic mean shift clustering algorithms maintain a set of data points the same size as the input data set. Initially, this set is copied from the input set. Then this set is iteratively replaced by the mean of those points in the set that are within a given distance of that point. By contrast, k-means restricts this updated set to k points usually much less than the number of points in the input data set, and replaces each point in this set by the mean of all points in the input set that are closer to that point than any other (e.g. within the Voronoi partition of each updating point). A mean shift algorithm that is similar then to k-means, called likelihood mean shift, replaces the set of points undergoing replacement by the mean of all points in the input set that are within a given distance of the changing set.[34] One of the advantages of mean shift over k-means is that there is no need to choose the number of clusters, because mean shift is likely to find only a few clusters if indeed only a small number exist. However, mean shift can be much slower than k-means, and still requires selection of a bandwidth parameter. Mean shift has soft variants much as k-means does.
Principal component analysis (PCA)
It was asserted[35][36] that the relaxed solution of k-means clustering, specified by the cluster indicators, is given by principal component analysis (PCA), and the PCA subspace spanned by the principal directions is identical to the cluster centroid subspace. However, that PCA is a useful relaxation of k-means clustering was not a new result,[37] and it is straightforward to uncover counterexamples to the statement that the cluster centroid subspace is spanned by the principal directions.[38]
Independent component analysis (ICA)
It has been shown in [39] that under sparsity assumptions and when input data is pre-processed with the whitening transformation k-means produces the solution to the linear Independent component analysis task. This aids in explaining the successful application of k-means to feature learning.
Bilateral filtering
k-means implicitly assumes that the ordering of the input data set does not matter. The bilateral filter is similar to K-means and mean shift in that it maintains a set of data points that are iteratively replaced by means. However, the bilateral filter restricts the calculation of the (kernel weighted) mean to include only points that are close in the ordering of the input data.[34] This makes it applicable to problems such as image denoising, where the spatial arrangement of pixels in an image is of critical importance.
Similar problems
The set of squared error minimizing cluster functions also includes the k-medoids algorithm, an approach which forces the center point of each cluster to be one of the actual points, i.e., it uses medoids in place of centroids.
Software implementations
Free
- CrimeStat implements two spatial k-means algorithms, one of which allows the user to define the starting locations.
- ELKI contains k-means (with Lloyd and MacQueen iteration, along with different initializations such as k-means++ initialization) and various more advanced clustering algorithms.
- Julia contains a k-means implementation in the Clustering package.[40]
- Mahout contains a MapReduce based k-means.
- MLPACK contains a C++ implementation of k-means.
- Octave contains k-means.
- OpenCV contains a k-means implementation.
- R contains three k-means variations.[1][3][6]
- SciPy and scikit-learn contain multiple k-means implementations.
- Spark MLlib implements a distributed k-means algorithm.
- Torch contains an unsup package that provides k-means clustering.
- Weka contains k-means and x-means.
Commercial
See also
- Centroidal Voronoi tessellation
- k q-flats
- Linde–Buzo–Gray algorithm
- Self-organizing map
- Head/tail Breaks
References
- 1 2 MacQueen, J. B. (1967). Some Methods for classification and Analysis of Multivariate Observations. Proceedings of 5th Berkeley Symposium on Mathematical Statistics and Probability. University of California Press. pp. 281–297. MR 0214227. Zbl 0214.46201. Retrieved 2009-04-07.
- ↑ Steinhaus, H. (1957). "Sur la division des corps matériels en parties". Bull. Acad. Polon. Sci. (in French) 4 (12): 801–804. MR 0090073. Zbl 0079.16403.
- 1 2 Lloyd, S. P. (1957). "Least square quantization in PCM". Bell Telephone Laboratories Paper. Published in journal much later: Lloyd., S. P. (1982). "Least squares quantization in PCM" (PDF). IEEE Transactions on Information Theory 28 (2): 129–137. doi:10.1109/TIT.1982.1056489. Retrieved 2009-04-15.
- ↑ E.W. Forgy (1965). "Cluster analysis of multivariate data: efficiency versus interpretability of classifications". Biometrics 21: 768–769. JSTOR 2528559.
- ↑ J.A. Hartigan (1975). Clustering algorithms. John Wiley & Sons, Inc.
- 1 2 3 Hartigan, J. A.; Wong, M. A. (1979). "Algorithm AS 136: A K-Means Clustering Algorithm". Journal of the Royal Statistical Society, Series C 28 (1): 100–108. JSTOR 2346830.
- ↑ MacKay, David (2003). "Chapter 20. An Example Inference Task: Clustering" (PDF). Information Theory, Inference and Learning Algorithms. Cambridge University Press. pp. 284–292. ISBN 0-521-64298-1. MR 2012999.
- ↑ Since the square root is a monotone function, this also is the minimum Euclidean distance assignment.
- 1 2 Hamerly, G. and Elkan, C. (2002). "Alternatives to the k-means algorithm that find better clusterings" (PDF). Proceedings of the eleventh international conference on Information and knowledge management (CIKM).
- ↑ Vattani., A. (2011). "k-means requires exponentially many iterations even in the plane" (PDF). Discrete and Computational Geometry 45 (4): 596–616. doi:10.1007/s00454-011-9340-1.
- 1 2 Arthur, D.; Manthey, B.; Roeglin, H. (2009). "k-means has polynomial smoothed complexity". Proceedings of the 50th Symposium on Foundations of Computer Science (FOCS).
- ↑ Aloise, D.; Deshpande, A.; Hansen, P.; Popat, P. (2009). "NP-hardness of Euclidean sum-of-squares clustering". Machine Learning 75: 245–249. doi:10.1007/s10994-009-5103-0.
- ↑ Dasgupta, S. and Freund, Y. (July 2009). "Random Projection Trees for Vector Quantization". Information Theory, IEEE Transactions on 55: 3229–3242. arXiv:0805.1390. doi:10.1109/TIT.2009.2021326.
- ↑ Mahajan, M.; Nimbhorkar, P.; Varadarajan, K. (2009). "The Planar k-Means Problem is NP-Hard". Lecture Notes in Computer Science 5431: 274–285. doi:10.1007/978-3-642-00202-1_24.
- ↑ Inaba, M.; Katoh, N.; Imai, H. (1994). Applications of weighted Voronoi diagrams and randomization to variance-based k-clustering. Proceedings of 10th ACM Symposium on Computational Geometry. pp. 332–339. doi:10.1145/177424.178042.
- ↑ Arthur; Abhishek Bhowmick (2009). A theoretical analysis of Lloyd's algorithm for k-means clustering (PDF) (Thesis).
- 1 2 Phillips, Steven J. (2002-01-04). Mount, David M.; Stein, Clifford, eds. Acceleration of K-Means and Related Clustering Algorithms. Lecture Notes in Computer Science. Springer Berlin Heidelberg. pp. 166–177. doi:10.1007/3-540-45643-0_13. ISBN 978-3-540-43977-6.
- 1 2 Elkan, C. (2003). "Using the triangle inequality to accelerate k-means" (PDF). Proceedings of the Twentieth International Conference on Machine Learning (ICML).
- 1 2 Hamerly, Greg. "Making k-means even faster". citeseerx.ist.psu.edu. Retrieved 2015-12-10.
- ↑ Kanungo, T.; Mount, D. M.; Netanyahu, N. S.; Piatko, C. D.; Silverman, R.; Wu, A. Y. (2002). "An efficient k-means clustering algorithm: Analysis and implementation" (PDF). IEEE Trans. Pattern Analysis and Machine Intelligence 24: 881–892. doi:10.1109/TPAMI.2002.1017616. Retrieved 2009-04-24.
- ↑ Drake, Jonathan (2012). "Accelerated k-means with adaptive distance bounds" (PDF). the 5th NIPS Workshop on Optimization for Machine Learning, OPT2012.
- ↑ Dhillon, I. S.; Modha, D. M. (2001). "Concept decompositions for large sparse text data using clustering". Machine Learning 42 (1): 143–175. doi:10.1023/a:1007612920971.
- ↑ Amorim, R.C.; Mirkin, B. (2012). "Minkowski Metric, Feature Weighting and Anomalous Cluster Initialisation in K-Means Clustering". Pattern Recognition 45 (3): 1061–1075. doi:10.1016/j.patcog.2011.08.012.
- ↑ Amorim, R.C.; Hennig, C. (2015). "Recovering the number of clusters in data sets with noise features using feature rescaling factors". Information Sciences 324: 126–145. doi:10.1016/j.ins.2015.06.039.
- ↑ Mirkes, E.M. "K-means and K-medoids applet.". Retrieved 2 January 2016.
- ↑ Honarkhah, M; Caers, J (2010). "Stochastic Simulation of Patterns Using Distance-Based Pattern Modeling". Mathematical Geosciences 42: 487–517. doi:10.1007/s11004-010-9276-7.
- 1 2 3 Coates, Adam; Ng, Andrew Y. (2012). "Learning feature representations with k-means" (PDF). In G. Montavon, G. B. Orr, K.-R. Müller. Neural Networks: Tricks of the Trade. Springer.
- ↑ Csurka, Gabriella; Dance, Christopher C.; Fan, Lixin; Willamowski, Jutta; Bray, Cédric (2004). Visual categorization with bags of keypoints (PDF). ECCV Workshop on Statistical Learning in Computer Vision.
- 1 2 Coates, Adam; Lee, Honglak; Ng, Andrew Y. (2011). An analysis of single-layer networks in unsupervised feature learning (PDF). International Conference on Artificial Intelligence and Statistics (AISTATS).
- ↑ Schwenker, Friedhelm; Kestler, Hans A.; Palm, Günther (2001). "Three learning phases for radial-basis-function networks". Neural Networks 14: 439–458. doi:10.1016/s0893-6080(01)00027-2. CiteSeerX: 10
.1 ..1 .109 .312 - ↑ Lin, Dekang; Wu, Xiaoyun (2009). Phrase clustering for discriminative learning (PDF). Annual Meeting of the ACL and IJCNLP. pp. 1030–1038.
- ↑ Press, WH; Teukolsky, SA; Vetterling, WT; Flannery, BP (2007). "Section 16.1. Gaussian Mixture Models and k-Means Clustering". Numerical Recipes: The Art of Scientific Computing (3rd ed.). New York: Cambridge University Press. ISBN 978-0-521-88068-8.
- ↑ Aharon, Michal; Elad, Michael; Bruckstein, Alfred (2006). "K-SVD: An Algorithm for Designing Overcomplete Dictionaries for Sparse Representation" (PDF).
- 1 2 Little, M.A.; Jones, N.S. (2011). "Generalized Methods and Solvers for Piecewise Constant Signals: Part I" (PDF). Proceedings of the Royal Society A 467: 3088–3114. doi:10.1098/rspa.2010.0671.
- ↑ H. Zha, C. Ding, M. Gu, X. He and H.D. Simon (Dec 2001). "Spectral Relaxation for K-means Clustering" (PDF). Neural Information Processing Systems vol.14 (NIPS 2001) (Vancouver, Canada): 1057–1064.
- ↑ Chris Ding and Xiaofeng He (July 2004). "K-means Clustering via Principal Component Analysis" (PDF). Proc. of Int'l Conf. Machine Learning (ICML 2004): 225–232.
- ↑ Drineas, P.; A. Frieze; R. Kannan; S. Vempala; V. Vinay (2004). "Clustering large graphs via the singular value decomposition" (PDF). Machine learning 56: 9–33. doi:10.1023/b:mach.0000033113.59016.96. Retrieved 2012-08-02.
- ↑ Cohen, M.; S. Elder; C. Musco; C. Musco; M. Persu (2014). "Dimensionality reduction for k-means clustering and low rank approximation (Appendix B)". ArXiv. Retrieved 2014-11-29.
- ↑ Alon Vinnikov and Shai Shalev-Shwartz (2014). "K-means Recovers ICA Filters when Independent Components are Sparse" (PDF). Proc. of Int'l Conf. Machine Learning (ICML 2014).
- ↑ Clustering.jl www.github.com
- ↑ http://help.sap.com/saphelp_hanaplatform/helpdata/en/53/e6908794ce4bcaa440f5c4348f3d14/content.htm