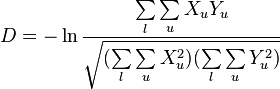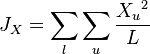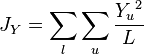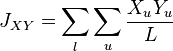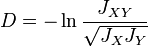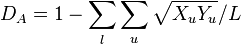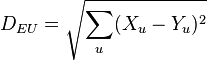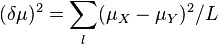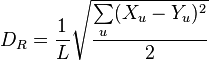Genetic distance
Genetic distance is a measure of the genetic divergence between species or between populations within a species.[1] Populations with many similar alleles have small genetic distances. This indicates that they are closely related and have a recent common ancestor.
Genetic distance is useful for reconstructing the history of populations. For example, evidence from genetic distance suggests that African and Eurasian people diverged about 100,000 years ago.[2] Genetic distance is also used for understanding the origin of biodiversity. For example, the genetic distances between different breeds of domesticated animals are often investigated in order to determine which breeds should be protected to maintain genetic diversity.[3]
Biological foundation
In the genome of an organism, each gene is located at a specific place called the locus for that gene. Allelic variations at these loci cause phenotypic variation within species (e.g. hair colour, eye colour). However, most alleles do not have an observable impact on the phenotype. Within a population new alleles generated by mutation either die out or spread throughout the population. When a population is split into different isolated populations (by either geographical or ecological factors), mutations that occur after the split will be present only in the isolated population. Random fluctuation of allele frequencies also produces genetic differentiation between populations. This process is known as genetic drift. By examining the differences between allele frequencies between the populations and computing genetic distance, we can estimate how long ago the two populations were separated.[4]
Measures of genetic distance
Although it is simple to define genetic distance as a measure of genetic divergence, there are several different statistical measures that have been proposed. This has happened because different authors considered different evolutionary models. The most commonly used are Nei's genetic distance,[4] Cavalli-Sforza and Edwards measure,[5] and Reynolds, Weir and Cockerham's genetic distance,[6] listed below.
In all the formulae in this section, 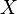 and
and 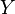 represent two different populations for which
represent two different populations for which  loci have been studied. Let
loci have been studied. Let 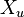 represent the
represent the 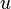 th allele at the
th allele at the 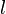 th locus.
th locus.
Nei's standard genetic distance
In 1972, Masatoshi Nei published what came to be known as Nei's standard genetic distance. This distance has the nice property that if the rate of genetic change (amino acid substitution) is constant per year or generation then Nei's standard genetic distance (D) increases in proportion to divergence time. This measure assumes that genetic differences are caused by mutation and genetic drift.[4]
This distance can also be expressed in terms of the arithmetic mean of gene identity. Let 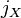 be the probability for the two members of population
be the probability for the two members of population 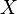 having the same allele at a particular locus and
having the same allele at a particular locus and 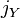 be the corresponding probability in population
be the corresponding probability in population 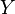 . Also, let
. Also, let 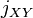 be the probability for a member of
be the probability for a member of 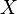 and a member of
and a member of 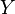 having the same allele. Now let
having the same allele. Now let 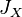 ,
, 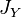 and
and 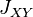 represent the arithmetic mean of
represent the arithmetic mean of 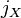 ,
, 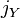 and
and 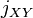 over all loci, respectively. In other words
over all loci, respectively. In other words
where  is the total number of loci examined.[7]
is the total number of loci examined.[7]
Nei's standard distance can then be written as[4]
Cavalli-Sforza chord distance
In 1967 Luigi Luca Cavalli-Sforza and A. W. F. Edwards published this measure. It assumes that genetic differences arise due to genetic drift only. One major advantage of this measure is that the populations are represented in a hypersphere, the scale of which is one unit per gene substitution. The chord distance in the hyperdimensional sphere is given by[1][5]
Some authors drop the factor 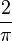 to simplify the formula at the cost of losing the property that the scale is one unit per gene substitution.
to simplify the formula at the cost of losing the property that the scale is one unit per gene substitution.
Reynolds, Weir, and Cockerham's genetic distance
In 1983, this measure was published by John Reynolds, B.S. Weir and C. Clark Cockerham.
This measure assumes that genetic differentiation occurs only by genetic drift without mutations. It estimates the coancestry coefficient 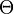 which provides a measure of the genetic divergence by:[6]
which provides a measure of the genetic divergence by:[6]
Other measures of genetic distance
Many other measures of genetic distance have been proposed with varying success.
Nei's DA distance 1983
This distance assumes that genetic differences arise due to mutation and genetic drift, but this distance measure is known to give more reliable population trees than other distances particularly for microsatellite DNA data.[8][9]
Euclidean distance
Goldstein distance 1995
It was specifically developed for microsatellite markers and is based on the stepwise-mutation model (SMM).  and
and  are the means of the allele sizes in population X and Y.[10]
are the means of the allele sizes in population X and Y.[10]
Nei's minimum genetic distance 1973
This measure assumes that genetic differences arise due to mutation and genetic drift.[2]
Roger's distance 1972
Fixation index
A commonly used measure of genetic distance is the fixation index which varies between 0 and 1. A value of 0 indicates that two populations are genetically identical whereas a value of 1 indicates that two populations are different species. No mutation is assumed.
Software
- PHYLIP uses GENDIST
- Nei's standard genetic distance 1972
- Cavalli-Sforza and Edwards 1967
- Reynolds, Weir, and Cockerham's 1983
- TFPGA
- Nei's standard genetic distance (original and unbiased)
- Nei's minimum genetic distance (original and unbiased)
- Wright's (1978) modification of Roger's (1972) distance
- Reynolds, Weir, and Cockerham's 1983
- GDA
- POPGENE
- POPTREE2 Takezaki, Nei, and Tamura (2010, 2014)
- Commonly used genetic distances and gene diversity analysis
- DISPAN
- Nei's standard genetic distance 1972
- Nei's DA distance between populations 1983
See also
- Coefficient of relationship
- Degree of consanguinity
- Human genetic variation
- Human genetic clustering
- Phylogenetics
- Allele frequency
References
- 1 2 3 Nei, M. (1987). Molecular Evolutionary Genetics. (Chapter 9). New York: Columbia University Press.
- 1 2 Nei, M. and A. K. Roychoudhury (1974). "Genic variation within and between the three major races of man, Caucasoids, Negroids, and Mongoloids". The American Journal of Human Genetics 26: 421–443. PMC 1762596. PMID 4841634.
- ↑ Ruane, J. (1999). A critical review of the value of genetic distance studies in conservation of animal genetic resources. Journal of Animal Breeding and Genetics, 116(5), 317-323. Chicago.
- 1 2 3 4 Nei, M. (1972). "Genetic distance between populations". Am. Nat. 106: 283–292. doi:10.1086/282771.
- 1 2 L.L. Cavalli-Sforza, A.W.F. Edwards (1967). "Phylogenetic Analysis -Models and Estimation Procedures". The American Journal of Human Genetics 19 (3 Part I (May)).
- 1 2 John Reynolds, B.S. Weir, C. Clark Cockerham (November 1983). "Estimation of the coancestry coefficient: Basis for a short-term genetic distance". Genetics 105: 767–779.
- ↑ Nei, M. (1987) Genetic distance and molecular phylogeny. In: Population Genetics and Fishery Management (N. Ryman and F. Utter, eds.), University of Washington Press, Seattle, WA, pp. 193-223.
- ↑ Nei, M., F. Tajima, & Y. Tateno (1983) Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 19:153-170.
- ↑ Takezaki, N. and Nei, M. (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389-399.
- ↑ Gillian Cooper; William Amos; Richard Bellamy; Mahveen Ruby Siddiqui; Angela Frodsham; Adrian V. S. Hill; David C. Rubinsztein (1999). "An Empirical Exploration of the
 Genetic Distance for 213 Human Microsatellite Markers". The American Journal of Human Genetics 65: 1125–1133. doi:10.1086/302574.
Genetic Distance for 213 Human Microsatellite Markers". The American Journal of Human Genetics 65: 1125–1133. doi:10.1086/302574.
- ↑ Rogers, J. S. (1972). Measures of similarity and genetic distance. In Studies in Genetics VII. pp. 145−153. University of Texas Publication 7213. Austin, Texas.
External links
- The Estimation of Genetic Distance and Population Substructure from Microsatellite allele frequency data., Brent W. Murray (May 1996), McMaster University website on genetic distance
- Computing distance by stepwise genetic distance model, web pages of Bruce Walsh at the Department of Ecology and Evolutionary Biology at the University of Arizona