Geminiviridae
| Geminiviridae | |
|---|---|
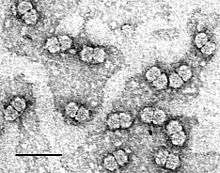 | |
| Purified Maize streak virus (MSV) particles stained with uranyl acetate. Size bar indicates 50 nm. | |
| Virus classification | |
| Group: | Group II (ssDNA) |
| Order: | Unassigned |
| Family: | Geminiviridae |
| Genera | |
Geminiviridae is a family of plant viruses. There are currently 325 species in this family, divided among 7 genera. Diseases associated with this family include: bright yellow mosaic, yellow mosaic, yellow mottle, leaf curling, stunting, streaks, reduced yields.[1][2] They have single-stranded circular DNA genomes encoding genes that diverge in both directions from a virion strand origin of replication (i.e. geminivirus genomes are ambisense). According to the Baltimore classification they are considered class II viruses. It is the largest known family of single stranded DNA viruses.
Mastrevirus transmission is via various leafhopper species (e.g. maize streak virus and other African streak viruses are transmitted by Cicadulina mbila), curtoviruses and the only known topocuvirus species, Tomato pseudo-curly top virus, are transmitted by treehopper species (e.g.Tomato pseudo-curly top virus is transmitted by the treehopper Micrutalis malleifera), and begomoviruses are transmitted by the whitefly species, Bemisia tabaci.
These viruses are responsible for a significant amount of crop damage worldwide. Epidemics of geminivirus diseases have arisen due to a number of factors, including the recombination of different geminiviruses coinfecting a plant, which enables novel, possibly virulent viruses to be developed. Other contributing factors include the transport of infected plant material to new locations, expansion of agriculture into new growing areas, and the expansion and migration of vectors that can spread the virus from one plant to another.[3]
Virology
The genome can either be a single component between 2500-3100 nucleotides, or, in the case of some begomoviruses, two similar-sized components each between 2600 and 2800 nucleotides. They have elongated, geminate capsids with two incomplete T=1 icosahedra joined at the missing vertex. The capsids range in size from 18-20 nm in diameter with a length of about 30 nm. Begomoviruses with two component (i.e. bipartite) genomes have these components separated into two different particles both of which must usually be transmitted together to initiate a new infection within a suitable host cell.
| Genus | Structure | Symmetry | Capsid | Genomic Arrangement | Genomic Segmentation |
|---|---|---|---|---|---|
| Eragrovirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Monopartite |
| Curtovirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Monopartite |
| Begomovirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Segmented |
| Becurtovirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Monopartite |
| Topocuvirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Monopartite |
| Turncurtovirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Monopartite |
| Mastrevirus | Twinned Icosahedral | Incomplete T=1 | Non-Enveloped | Circular | Monopartite |
Taxonomy
Group: ssDNA
- Family: Geminiviridae
- Genus: Becurtovirus
- Beet curly top Iran virus
- Spinach curly top Arizona virus
- Genus: Begomovirus
- Abutilon Brazil virus
- Abutilon mosaic Bolivia virus
- Abutilon mosaic Brazil virus
- Abutilon mosaic virus
- African cassava mosaic virus
- Ageratum enation virus
- Ageratum leaf curl Cameroon virus
- Ageratum leaf curl virus
- Ageratum yellow vein Hualian virus
- Ageratum yellow vein Sri Lanka virus
- Ageratum yellow vein virus
- Allamanda leaf curl virus
- Alternanthera yellow vein virus
- Bean calico mosaic virus
- Bean chlorosis virus
- Bean dwarf mosaic virus
- Bean golden mosaic virus
- Bean golden yellow mosaic virus
- Bean yellow mosaic Mexico virus
- Bhendi yellow vein Bhubhaneswar virus
- Bhendi yellow vein Haryana virus
- Bhendi yellow vein India virus
- Bhendi yellow vein mosaic virus
- Bitter gourd yellow vein virus
- Blainvillea yellow spot virus
- Blechum interveinal chlorosis virus
- Boerhavia yellow spot virus
- Cabbage leaf curl Jamaica virus
- Cabbage leaf curl virus
- Centrosema yellow spot virus
- Chayote yellow mosaic virus
- Chilli leaf curl virus
- Chino del tomate Amazonas virus
- Chino del tomate virus
- Cleome golden mosaic virus
- Cleome leaf crumple virus
- Clerodendron golden mosaic virus
- Corchorus golden mosaic virus
- Corchorus yellow spot virus
- Corchorus yellow vein virus
- Cotton leaf crumple virus
- Cotton leaf curl Alabad virus
- Cotton leaf curl Bangalore virus
- Cotton leaf curl Gezira virus
- Cotton leaf curl Kokhran virus
- Cotton leaf curl Multan virus
- Cowpea golden mosaic virus
- Croton yellow vein mosaic virus
- Cucurbit leaf crumple virus
- Dalechampia chlorotic mosaic virus
- Datura leaf distortion virus
- Desmodium leaf distortion virus
- Dicliptera yellow mottle Cuba virus
- Dicliptera yellow mottle virus
- Dolichos yellow mosaic virus
- East African cassava mosaic Cameroon virus
- East African cassava mosaic Kenya virus
- East African cassava mosaic Malawi virus
- East African cassava mosaic virus
- East African cassava mosaic Zanzibar virus
- Eclipta yellow vein virus
- Erectites yellow mosaic virus
- Eupatorium yellow vein mosaic virus
- Eupatorium yellow vein virus
- Euphorbia leaf curl Guangxi virus
- Euphorbia leaf curl virus
- Euphorbia mosaic virus
- Euphorbia yellow mosaic virus
- Hollyhock leaf crumple virus
- Hollyhock leaf curl virus
- Honeysuckle yellow vein Kagoshima virus
- Honeysuckle yellow vein mosaic virus
- Honeysuckle yellow vein virus
- Horsegram yellow mosaic virus
- Indian cassava mosaic virus
- Jacquemontia mosaic Yucatan virus
- Jatropha mosaic India virus
- Kenaf leaf curl virus
- Kudzu mosaic virus
- Leonurus mosaic virus
- Lindernia anagallis yellow vein virus
- Ludwigia yellow vein Vietnam virus
- Ludwigia yellow vein virus
- Luffa yellow mosaic virus
- Macroptilium golden mosaic virus
- Macroptilium mosaic Puerto Rico virus
- Macroptilium yellow mosaic Florida virus
- Macroptilium yellow mosaic virus
- Macroptilium yellow net virus
- Macroptilium yellow spot virus
- Macroptilium yellow vein virus
- Malvastrum leaf curl Guangdong virus
- Malvastrum leaf curl virus
- Malvastrum yellow leaf curl virus
- Malvastrum yellow mosaic Helshire virus
- Malvastrum yellow mosaic Jamaica virus
- Malvastrum yellow mosaic virus
- Malvastrum yellow vein Changa Manga virus
- Malvastrum yellow vein Honghe virus
- Malvastrum yellow vein virus
- Malvastrum yellow vein Yunnan virus
- Melon chlorotic leaf curl virus
- Melon chlorotic mosaic virus
- Merremia mosaic Puerto Rico virus
- Merremia mosaic virus
- Mesta yellow vein mosaic virus
- Mimosa yellow leaf curl virus
- Mungbean yellow mosaic India virus
- Mungbean yellow mosaic virus
- Okra enation leaf curl virus
- Okra leaf curl Cameroon virus
- Okra mottle virus
- Okra yellow crinkle virus
- Okra yellow mosaic Mexico virus
- Okra yellow mottle Iguala virus
- Okra yellow vein mosaic virus
- Papaya leaf crumple virus
- Papaya leaf curl China virus
- Papaya leaf curl Guandong virus
- Papaya leaf curl virus
- Passionfruit severe leaf distortion virus
- Pedilenthus leaf curl virus
- Pepper golden mosaic virus
- Pepper huasteco yellow vein virus
- Pepper leaf curl Bangladesh virus
- Pepper leaf curl Lahore virus
- Pepper leaf curl virus
- Pepper leaf curl Yunnan virus
- Pepper yellow leaf curl Indonesia virus
- Pepper yellow vein Mali virus
- Potato yellow mosaic Panama virus
- Potato yellow mosaic virus
- Pumpkin yellow mosaic virus
- Radish leaf curl virus
- Rhynchosia golden mosaic Havana virus
- Rhynchosia golden mosaic Sinaloa virus
- Rhynchosia golden mosaic virus
- Rhynchosia golden mosaic Yucatan virus
- Rhynchosia mild mosaic virus
- Rhynchosia rugose golden mosaic virus
- Rhynchosia yellow mosaic virus
- Rose leaf curl virus
- Senecio yellow mosaic virus
- Sida golden mosaic Braco virus
- Sida golden mosaic Buckup virus
- Sida golden mosaic Costa Rica virus
- Sida golden mosaic Florida virus
- Sida golden mosaic Honduras virus
- Sida golden mosaic Liguanea virus
- Sida golden mosaic virus
- Sida golden mottle virus
- Sida golden yellow vein virus
- Sida leaf curl virus
- Sida micrantha mosaic virus
- Sida mosaic Alagoas virus
- Sida mosaic Bolivia virus 1
- Sida mosaic Bolivia virus 2
- Sida mosaic Sinaloa virus
- Sida mottle Alagoas virus
- Sida mottle virus
- Sida yellow blotch virus
- Sida yellow mosaic Alagoas virus
- Sida yellow mosaic China virus
- Sida yellow mosaic virus
- Sida yellow mosaic Yucatan virus
- Sida yellow mottle virus
- Sida yellow net virus
- Sida yellow vein Madurai virus
- Sida yellow vein Vietnam virus
- Sida yellow vein virus
- Siegesbeckia yellow vein Guangxi virus
- Siegesbeckia yellow vein virus
- South African cassava mosaic virus
- Soybean blistering mosaic virus
- Soybean chlorotic spot virus
- Soybean crinkle leaf virus
- Soybean mild mottle virus
- Spilanthes yellow vein virus
- Squash leaf curl China virus
- Squash leaf curl Philippines virus
- Squash leaf curl virus
- Squash leaf curl Yunnan virus
- Squash mild leaf curl virus
- Sri Lankan cassava mosaic virus
- Stachytarpheta leaf curl virus
- Sweet potato leaf curl Canary virus
- Sweet potato leaf curl China virus
- Sweet potato leaf curl Georgia virus
- Sweet potato leaf curl Sao Paulo virus
- Sweet potato leaf curl South Carolina virus
- Sweet potato leaf curl Uganda virus
- Sweet potato leaf curl virus
- Sweet potato mosaic virus
- Tobacco curly shoot virus
- Tobacco leaf curl Cuba virus
- Tobacco leaf curl Japan virus
- Tobacco leaf curl Pusa virus
- Tobacco leaf curl Thailand virus
- Tobacco leaf curl Yunnan virus
- Tobacco leaf curl Zimbabwe virus
- Tobacco leaf rugose virus
- Tobacco mottle leaf curl virus
- Tobacco yellow crinkle virus
- Tomato chino La Paz virus
- Tomato chlorotic leaf distortion virus
- Tomato chlorotic mottle virus
- Tomato common mosaic virus
- Tomato curly stunt virus
- Tomato dwarf leaf virus
- Tomato golden mosaic virus
- Tomato golden mottle virus
- Tomato golden vein virus
- Tomato leaf curl Anjouan virus
- Tomato leaf curl Arusha virus
- Tomato leaf curl Bangalore virus
- Tomato leaf curl Bangladesh virus
- Tomato leaf curl Cebu virus
- Tomato leaf curl China virus
- Tomato leaf curl Comoros virus
- Tomato leaf curl Diana virus
- Tomato leaf curl Ghana virus
- Tomato leaf curl Guangdong virus
- Tomato leaf curl Guangxi virus
- Tomato leaf curl Gujarat virus
- Tomato leaf curl Hainan virus
- Tomato leaf curl Hanoi virus
- Tomato leaf curl Hsinchu virus
- Tomato leaf curl Iran virus
- Tomato leaf curl Java virus
- Tomato leaf curl Joydebpur virus
- Tomato leaf curl Karnataka virus
- Tomato leaf curl Kerala virus
- Tomato leaf curl Kumasi virus
- Tomato leaf curl Laos virus
- Tomato leaf curl Madagascar virus
- Tomato leaf curl Malaysia virus
- Tomato leaf curl Mali virus
- Tomato leaf curl Mindanao virus
- Tomato leaf curl Moheli virus
- Tomato leaf curl Namakely virus
- Tomato leaf curl New Delhi virus
- Tomato leaf curl Nigeria virus
- Tomato leaf curl Oman virus
- Tomato leaf curl Philippines virus
- Tomato leaf curl Pune virus
- Tomato leaf curl Seychelles virus
- Tomato leaf curl Sinaloa virus
- Tomato leaf curl Sri Lanka virus
- Tomato leaf curl Sudan virus
- Tomato leaf curl Taiwan virus
- Tomato leaf curl Toliara virus
- Tomato leaf curl Uganda virus
- Tomato leaf curl Vietnam virus
- Tomato leaf curl virus
- Tomato leaf deformation virus
- Tomato leaf distortion virus
- Tomato mild mosaic virus
- Tomato mild yellow leaf curl Aragua virus
- Tomato mosaic Havana virus
- Tomato mosaic leaf curl virus
- Tomato mottle leaf curl virus
- Tomato mottle Taino virus
- Tomato mottle virus
- Tomato rugose mosaic virus
- Tomato rugose yellow leaf curl virus
- Tomato severe leaf curl virus
- Tomato severe rugose virus
- Tomato yellow leaf curl Axarquia virus
- Tomato yellow leaf curl China virus
- Tomato yellow leaf curl Guangdong virus
- Tomato yellow leaf curl Indonesia virus
- Tomato yellow leaf curl Kanchanaburi virus
- Tomato yellow leaf curl Malaga virus
- Tomato yellow leaf curl Mali virus
- Tomato yellow leaf curl Sardinia virus
- Tomato yellow leaf curl Thailand virus
- Tomato yellow leaf curl Vietnam virus
- Tomato yellow leaf curl virus
- Tomato yellow leaf distortion virus
- Tomato yellow margin leaf curl virus
- Tomato yellow mottle virus
- Tomato yellow spot virus
- Tomato yellow vein streak virus
- Vernonia yellow vein virus
- Watermelon chlorotic stunt virus
- West African Asystasia virus 1
- West African Asystasia virus 2
- Wissadula golden mosaic virus
- Genus: Curtovirus
- Beet curly top virus
- Horseradish curly top virus
- Spinach severe curly top virus
- Genus: Mastrevirus
- Bromus catharticus striate mosaic virus
- Chickpea chlorosis Australia virus
- Chickpea chlorosis virus
- Chickpea chlorotic dwarf virus
- Chickpea redleaf virus
- Chickpea yellows virus
- Chloris striate mosaic virus
- Digitaria ciliaris striate mosaic virus
- Digitaria didactyla striate mosaic virus
- Digitaria streak virus
- Eragrostis minor streak virus
- Eragrostis streak virus
- Maize streak Reunion virus
- Maize streak virus
- Miscanthus streak virus
- Oat dwarf virus
- Panicum streak virus
- Paspalum dilatatum striate mosaic virus
- Paspalum striate mosaic virus
- Saccharum streak virus
- Sporolobus striate mosaic virus 1
- Sporolobus striate mosaic virus 2
- Sugarcane streak Egypt virus
- Sugarcane streak Reunion virus
- Sugarcane streak virus
- Tobacco yellow dwarf virus
- Urochloa streak virus
- Wheat dwarf India virus
- Wheat dwarf virus
Several additional genera have been proposed: Baminivirus, Nimivirus and Niminivirus.[4]
Some viruses have yet to be assigned a genus: one of these is the Grapevine Cabernet Franc-associated virus/Grapevine red blotch-associated virus/Grapevine redleaf-associated virus.[5]
Replication

Geminivirus genomes encode only a few proteins; thus, they are dependent on host cell factors for replication: these include factors such as DNA polymerase - and probably repair polymerases - in order to amplify their genomes, as well as transcription factors. Geminiviruses replicate via a rolling circle mechanism like bacteriophages such as M13, and many plasmids. Replication occurs within the nucleus of an infected plant cell. First the single-stranded circular DNA is converted to a double-stranded circular intermediate. This step involves the use of cellular DNA repair enzymes to produce a complementary negative-sense strand, using the viral genomic or plus-sense DNA strand as a template. The next step is the rolling circle phase, where the viral strand is cleaved at a specific site situated within the origin of replication by the viral Rep protein in order to initiate replication.[6] This process in a eukaryotic nucleus can give rise to concatemeric double-stranded forms of replicative intermediate genomes, although double-stranded unit circles can be isolated from infected plants and cells. New single-stranded DNA forms of the virus genome (plus-sense) are probably formed by interaction of the coat protein with replicating DNA intermediates, as genomes lacking a CP gene do not form ssDNA. The ssDNA is packaged into germinate particles in the nucleus. It is not clear if these particles can then leave the nucleus and be transmitted to surrounding cells as virions, or whether ssDNA associated with coat protein and a movement protein is the form of the genome that gets trafficked from cell to cell via the plasmodesmata.[7]
These viruses tend to be introduced into and initially infect differentiated plant cells, via the piercing mouthparts of the vector insect: however, these cells generally lack the host enzymes necessary for DNA replication, making it difficult for the virus to replicate. To overcome this block geminiviruses can induce plant cells to reenter the cell cycle from a quiescent state so that viral replication can occur.[8]
| Genus | Host Details | Tissue Tropism | Entry Details | Release Details | Replication Site | Assembly Site | Transmission |
|---|---|---|---|---|---|---|---|
| Eragrovirus | Plants | None | Viral movement; mechanical inoculation | Budding | Nucleus | Nucleus | Treehopper; leafhopper |
| Curtovirus | Dicotyledonous plants | Phloem-limited | Viral movement; mechanical inoculation | Budding | Nucleus | Nucleus | Beet leefhopper |
| Begomovirus | Dicotyledonous plants | Phloem; sieve; phloem-limited | Viral movement; mechanical inoculation | Budding | Nucleus | Nucleus | Bemisia tabaci whiteflies |
| Becurtovirus | Spinach | Phloem; sieve; phloem-limited | Viral movement; mechanical inoculation | Budding | Nucleus | Nucleus | Viral movement; contact |
| Topocuvirus | Dicotyledonous plants | None | Cell receptor endocytosis | Budding | Nucleus | Nucleus | Leafhopper |
| Turncurtovirus | Turnip | None | Cell receptor endocytosis | Budding | Nucleus | Nucleus | Leafhopper |
| Mastrevirus | Monocots[9] | None | Viral movement; mechanical inoculation | Budding | Nucleus | Nucleus | Leafhopper |
Evolution
These viruses may have evolved from a phytoplasma plasmid.[10] Geminiviruses are capable of horizontal gene transfer of genetic information to the plant host.[11]
References
- ↑ "Viral Zone". ExPASy. Retrieved 15 June 2015.
- 1 2 ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- ↑ Gray and Banerjee; Banerjee, N (1999). "Mechanisms of Arthropod Transmission of Plant and Animal Viruses". Microbiol Mol Biol Rev. 63 (1): 128–148. PMC 98959. PMID 10066833.
- ↑ Ng TF, Marine R, Wang C, Simmonds P, Kapusinszky B, Bodhidatta L, Oderinde BS, Wommack KE, Delwart E (2012) High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol
- ↑ Poojari S, Alabi OJ, Fofanov VY, Naidu RA (2013) A leafhopper-transmissible DNA virus with novel evolutionary lineage in the family geminiviridae implicated in grapevine redleaf disease by next-generation sequencing" PLoS One 8(6) e64194. doi:10.1371/journal.pone.0064194
- ↑ Chasan R (1995). "Geminiviruses: A Twin Approach to Replication" (PDF). Plant Cell 7 (6): 659–661. doi:10.1105/tpc.7.6.659.
- ↑ Gutierrez C (2000). "NEW EMBO MEMBERS' REVIEW: DNA replication and cell cycle in plants: learning from geminiviruses". EMBO 19 (5): 792–799. doi:10.1093/emboj/19.5.792. PMC 305619. PMID 10698921.
- ↑ Hanley Bowdoin lab
- ↑ http://viralzone.expasy.org/all_by_species/110.html
- ↑ Krupovic M, Ravantti JJ, Bamford DH (2009). "Geminiviruses: a tale of a plasmid becoming a virus" (PDF). BMC Evol Biol 9: 112. doi:10.1186/1471-2148-9-112. PMC 2702318. PMID 19460138.
- ↑ Bejarano,E.R., Khashoggi,A.M., Witty,M. and Lichtenstein,C.P. (1994). Discovery of ancient recombination between geminiviral DNA and the nuclear genome of Nicotiana sp. Proceedings of the National Academy of Sciences 93:759-764.
External links
- Description of Plant Viruses
- MicrobiologyBytes: Plant Viruses
- International Committee on Taxonomy of Viruses
- Viralzone: Geminiviridae
- ICTV