Estrada index
In chemical graph theory, the Estrada index is a topological index of protein folding. The index was first defined by Ernesto Estrada as a measure of the degree of folding of a protein,[1] which is represented as a path-graph weighted by the dihedral or torsional angles of the protein backbone. This index of degree of folding has found multiple applications in the study of protein functions and protein-ligand interactions.
The name of this index as the “Estrada index” was proposed by de la Peña et al. in 2007.[2]
Derivation
Let  be a graph of size
be a graph of size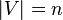 and let
and let 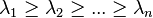 be a non-increasing ordering of the eigenvalues of its adjacency matrix
be a non-increasing ordering of the eigenvalues of its adjacency matrix 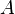 . The Estrada index is defined as
. The Estrada index is defined as
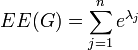
For a general graph, the index can be obtained as the sum of the subgraph centralities of all nodes in the graph. The subgraph centrality of node  is defined as[3]
is defined as[3]
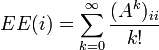
The subgraph centrality has the following closed form[3]
![EE(i)=(e^A)_{ii}=\sum_{j=1}^{n}[\varphi(i)]^2 e^{\lambda _j}](../I/m/aa5183ab1dc2ef899470255f61e93d09.png)
where 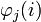 is the
is the  th entry of the
th entry of the 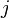 th eigenvector associated with the eigenvalue
th eigenvector associated with the eigenvalue  . It is straightforward to realise that[3]
. It is straightforward to realise that[3]
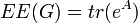
References
- ↑ Estrada, E. (2000). "Characterization of 3D molecular structure". Chem. Phys. Lett. (319): 713. Bibcode:2000CPL...319..713E. doi:10.1016/S0009-2614(00)00158-5.
- ↑ de la Peña, J. A.; Gutman, I.; Rada, J. (2007). "Estimating the Estrada index". Linear Algebra Appl. 427: 70–76. doi:10.1016/j.laa.2007.06.020.
- 1 2 3 Estrada, E.; Rodríguez-Velázquez, J.A. (2005). "Subgraph centrality in complex networks". Phys. Rev. E 71 (5): 056103. arXiv:cond-mat/0504730. Bibcode:2005PhRvE..71e6103E. doi:10.1103/PhysRevE.71.056103.
- Zhou, Bo; Gutman, Ivan (2009). "More on the Laplacian Estrada Index". Applic. Anal. Discr. Math. 3 (2): 371–378. doi:10.2298/AADM0902371Z.