DNA-directed RNA interference
DNA-directed RNA interference (ddRNAi) is a gene-silencing technique that utilizes DNA constructs to activate an animal cell’s endogenous RNA interference (RNAi) pathways. DNA constructs are designed to express self-complementary double-stranded RNAs, typically short-hairpin RNAs (shRNA), that once processed bring about silencing of a target gene or genes.[1] Any RNA, including endogenous mRNAs or viral RNAs, can be silenced by designing constructs to express double-stranded RNA complementary to the desired mRNA target.
This mechanism has great potential as a novel therapeutic to silence disease-causing genes. Proof-of-concept has been demonstrated across a range of disease models, including viral diseases such as HIV,[2] hepatitis B[3] or hepatitis C,[4] or diseases associated with altered expression of endogenous genes such as drug-resistant lung cancer,[5] neuropathic pain,[6] advanced cancer [7] and retinitis pigmentosa.[8]
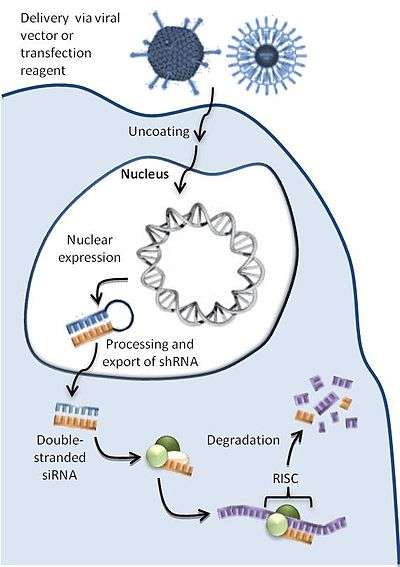
ddRNAi mechanism
As seen in Figure 1, a ddRNAi construct encoding an shRNA is packaged into a delivery vector or reagent tailored to target specific cells. Inside the cell, the DNA is transported to the nucleus where transcription machinery continually manufactures the encoded RNAs. The shRNA molecules are then processed by endogenous systems and enter the RNAi pathway and silence the desired genes.[1]
Long-term activity
Unlike small interfering RNA (siRNA) therapeutics that turn over within a cell and consequently only silence genes transiently, DNA constructs are continually transcribed, replenishing the cellular ‘dose’ of shRNA, thereby enabling long-term silencing of targeted genes. The ddRNAi mechanism, therefore, offers the potential for ongoing clinical benefit with reduced medical intervention.[2][4]
Organization of ddRNAi constructs
Figure 2 illustrates the most common type of ddRNAi DNA construct, which is designed to express a shRNA. This consists of a promoter sequence, driving expression of sense and antisense sequences separated by a loop sequence, followed by a transcriptional terminator. The antisense species processed from the shRNA can bind to the target RNA and specify its degradation. shRNA constructs typically encode sense and antisense sequences of 20 – 30 nucleotides. Flexibility in construct design is possible: for example, the positions of sense and antisense sequences can be reversed, and other modifications and additions can alter intracellular shRNA processing.[9] Moreover, a variety of promoter loop and terminator sequences can be used.[4]
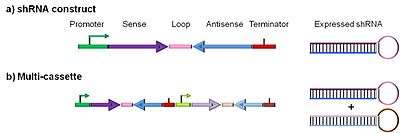
A particularly useful variant is a multi-cassette (Figure 2b). Designed to express two or more shRNAs, they can target multiple sequences for degradation simultaneously. This is a particularly useful strategy for targeting viruses. Natural sequence variations can render a single shRNA-target site unrecognizable preventing RNA degradation. Multi-cassette constructs that target multiple sites within the same viral RNA circumvent this issue.[4]
Delivery
Delivery of ddRNAi DNA constructs is simplified by the existence of a number of clinically-approved and well-characterized gene therapy vectors developed for the purpose. Delivery is a major challenge for RNAi-based therapeutics with new modifications and reagents continually being developed to optimize target cell delivery. Two broad strategies to facilitate delivery of DNA constructs to the desired cells are available: these use either viral vectors or one of a number of classes of transfection reagents.
In vivo delivery of ddRNAi constructs has been demonstrated using a range of vectors and reagents with different routes of administration (ROA).[3][4][6][8]
ddRNAi constructs have also been successfully delivered into host cells ex vivo, and then transplanted back into the host.[2][7][10]
For example, in a Phase I clinical trial at the City of Hope National Medical Center, California, US, four HIV-positive patients with non-Hodgkin’s lymphoma were successfully treated with autologous hematopoietic progenitor cells pre-transduced ex vivo with ddRNAi constructs using lentiviral vectors. This construct was designed to express three therapeutic RNAs, one of which was a shRNA, thereby combating HIV replication in three different ways:[2]
- shRNA, that silences the tat and rev genes of the HIV genome
- CCR5 ribozyme, inhibiting viral cell entry
- TAR decoy RNA, inhibiting initiation of viral transcription.
Ongoing expression of the shRNA has been confirmed in T cells, monocytes and B cells more than one year after transplantation.[2]
Therapeutic applications
Benitec Biopharma Ltd, through an exclusive license agreement with CSIRO, holds significant intellectual property for clinical applications of ddRNAi in humans and is pursuing programs in:[11]
- Neuropathic pain including cancer-associated pain, in association with the University of Queensland, QLD, Australia, and Stanford University, California, US
- Drug-resistant lung cancer, in association with the University of New South Wales, NSW, Australia
- Hepatitis B viral infection, now wholly owned by Benitec
- Hepatitis C viral infection, through its subsidiary Tacere Therapeutics Inc[12]
- Oculopharyngeal muscular dystrophy (OPMD), in association with the Royal Holloway, University of London, UK.
Further clinical research is being conducted through licensing agreements:[11]
- Retinitis pigmentosa, licensed exclusively to Genable Technologies, Dublin, Ireland[13]
- HIV/AIDS, licensed non-exclusively to Calimmune, Arizona, US[14]
- Huntington's disease, licensed exclusively to UniQure, Amsterdam, Netherlands.[15]
Neuropathic pain
Nervana is an investigational ddRNAi construct that knocks down the expression of protein kinase C gamma (PKCγ) known to be associated with neuropathic pain and morphine tolerance.[6][16]
Two conserved PKCγ sequences found across all key model species and humans have been identified, and both single and double DNA cassettes designed. In vitro, expression of PKCγ was silenced by 80%. When similar ddRNAi constructs were delivered intrathecally using a lentiviral vector, pain relief in a neuropathic-rat model was demonstrated.[6]
Drug-resistant non-small-cell lung cancer
The development of resistance to chemotherapies such as paclitaxel and cisplatin in non-small-cell lung cancer (NSCLC) is strongly associated with over expression of beta III tubulin. Investigations by the Children’s Cancer Institute Australia (University of NSW, Lowy Cancer Research Centre) demonstrated that beta III-tubulin knockdown by ddRNAi delayed tumor growth and increased chemosensitivity in mouse models.[5]
Tribetarna is a triple DNA cassette expressing three shRNA molecules that each separately target beta III tubulin and strongly inhibit its expression. Studies in an orthotopic-mouse model, where the construct is delivered by a modified polyethylenimine vector, jetPEI, that targets lung tissue are in progress.[16]
Hepatitis B viral infection
The hepatitis B virus (HBV) genome encodes its own DNA polymerase for replication. Biomics Biotechnologies has evaluated around 5000 siRNA sequences of this gene for effective knockdown; five sequences were chosen for further investigation and shown to have potent silencing activity when converted into shRNA expression cassettes. A multi-cassette construct, Hepbarna, is under preclinical development for delivery by an adeno-associated virus 8 (AAV-8) liver-targeting vector.[16]
Hepatitis C viral infection
Tacere Therapeutics Inc (now a subsidiary of Benitec Biopharma[12]) and Pfizer collaborated in the development of DNA construct TT-034 (also known as PF-05095808), an AAV-packaged triple cassette expressing three shRNA molecules targeting different areas of the hepatitis C viral (HCV) genome.[17] Preclinical testing has demonstrated the safety of this approach[4] and first-in-class human trials with this vector will commence in late 2013.
Oculopharyngeal muscular dystrophy
Classified as an orphan disease, there is currently no therapy for OPMD, caused by a mutation in the poly(A) binding protein nuclear 1 (PABPN1) gene. Silencing the mutant gene using ddRNAi offers a potential therapeutic approach.[16][18]
The Royal Holloway, UK, and The Institut de Myologie, Paris, in collaboration with Benitec Biopharma, is developing Pabparna, a ddRNAi construct for the treatment of OPMD. Construct design is under development and will be delivered either in vivo using an AAV-8 vector or ex vivo using a lentiviral vector.[16]
HIV/AIDS
Besides the ex vivo approach by the City of Hope National Medical Center discussed above, the Center for Infection and Immunity Amsterdam (CINIMA), University of Amsterdam, the Netherlands, is extensively researching the composition of multi-cassette DNA constructs to tackle HIV.[19]
Calimmune also has a non-exclusive license with Benitec Biopharma to pursue a novel ddRNAi therapeutic targeting up to three key viral and cellular genes against HIV.[10][14] The target genes and siRNA sequences have been identified and a clinical trial utilizing a ddRNAi construct has been designed.[20]
Safety concerns
As with all gene therapies, a number of safety and toxicity issues need to be evaluated during the development of ddRNAi therapeutics:
- Oncogene activation by viral insertion: Some gene therapy vectors integrate into the host genome, thereby acting as insertional mutagens. This was a particular issue with early retroviral vectors where insertions adjacent to oncogenes resulted in the development of lymphoid tumors.[21] AAV vectors are considered a low risk for host-genome integration, as adeno-associated virus infection has not been associated with the induction of cancers in humans despite widespread prevalence across the general population. Moreover, extensive clinical use of AAV vectors has provided no evidence of carcinogenicity. While lentiviral vectors do integrate into the genome they do not appear to show a propensity to activate oncogene expression.[22]
- Immune response to gene therapy vectors: An immunological response to an adenoviral vector resulted in the death of a patient in an early human trial. Careful monitoring of potential toxicities in preclinical testing and analyses of pre-existing antibodies to gene therapy vectors in patients minimizes such risks.
- Innate immune response: siRNAs have been shown to activate immune responses through interaction with Toll-like receptors leading to interferon responses. These receptors reside on the cells surface and so ddRNAi constructs – delivered directly into intracellular space – are not expected to induce this response.[23]
- Toxic effects due to over-expression of shRNAs: High level expression of shRNAs has been shown to be toxic. Strategies to minimize levels of shRNA expression[4] or promote precise processing of shRNAs[24] can overcome this problem.
- Off-target effects: Unintended silencing of genes that share sequence homology with expressed shRNAs can theoretically occur.[25] Careful selection of shRNA sequences and thorough preclinical testing of constructs can circumvent this issue.
Further reading
- Rice, R. R.; Muirhead, A. N.; Harrison, B. T.; Kassianos, A. J.; Sedlak, P. L.; Maugeri, N. J.; Goss, P. J.; Davey, J. R.; James, D. E.; Graham, M. W. (2005). "Simple, Robust Strategies for Generating DNA-Directed RNA Interference Constructs". Methods in enzymology. Methods in Enzymology 392: 405–419. doi:10.1016/S0076-6879(04)92024-1. ISBN 9780121827977. PMID 15644195.
- Liu, Y. P.; Westerink, J. T.; Brake, O.; Berkhout, B. (2011). "RNAi-Inducing Lentiviral Vectors for Anti-HIV-1 Gene Therapy". Antiviral RNAi. Methods in Molecular Biology 721. pp. 293–311. doi:10.1007/978-1-61779-037-9_18. ISBN 978-1-61779-036-2. PMID 21431693.
References
- 1 2 Lambeth, L. S.; Smith, C. A. (2013). "Short Hairpin RNA-Mediated Gene Silencing". SiRNA Design. Methods in Molecular Biology 942. pp. 205–232. doi:10.1007/978-1-62703-119-6_12. ISBN 978-1-62703-118-9. PMID 23027054.
- 1 2 3 4 5 Digiusto, D. L.; Krishnan, A.; Li, L.; Li, H.; Li, S.; Rao, A.; Mi, S.; Yam, P.; Stinson, S.; Kalos, M.; Alvarnas, J.; Lacey, S. F.; Yee, J. -K.; Li, M.; Couture, L.; Hsu, D.; Forman, S. J.; Rossi, J. J.; Zaia, J. A. (2010). "RNA-Based Gene Therapy for HIV with Lentiviral Vector-Modified CD34+ Cells in Patients Undergoing Transplantation for AIDS-Related Lymphoma". Science Translational Medicine 2 (36): 36ra43. doi:10.1126/scitranslmed.3000931. PMC 3130552. PMID 20555022.
- 1 2 Chen, C. -C.; Chang, C. -M.; Sun, C. -P.; Yu, C. -P.; Wu, P. -Y.; Jeng, K. -S.; Hu, C. -P.; Chen, P. -J.; Wu, J. -C.; Shih, C. H.; Gershwin, M. E.; Tao, M. -H. (2011). "Use of RNA interference to modulate liver adenoma development in a murine model transgenic for hepatitis B virus". Gene Therapy 19 (1): 25–33. doi:10.1038/gt.2011.60. PMID 21562593.
- 1 2 3 4 5 6 7 Suhy, D. A.; Kao, S. C.; Mao, T.; Whiteley, L.; Denise, H.; Souberbielle, B.; Burdick, A. D.; Hayes, K.; Wright, J. F.; Lavender, H.; Roelvink, P.; Kolykhalov, A.; Brady, K.; Moschos, S. A.; Hauck, B.; Zelenaia, O.; Zhou, S.; Scribner, C.; High, K. A.; Renison, S. H.; Corbau, R. (2012). "Safe, Long-term Hepatic Expression of Anti-HCV shRNA in a Nonhuman Primate Model". Molecular Therapy 20 (9): 1737–1749. doi:10.1038/mt.2012.119. PMC 3437581. PMID 22735378.
- 1 2 McCarroll, J. A.; Gan, P. P.; Liu, M.; Kavallaris, M. (2010). "III-Tubulin is a Multifunctional Protein Involved in Drug Sensitivity and Tumorigenesis in Non-Small Cell Lung Cancer". Cancer Research 70 (12): 4995–5003. doi:10.1158/0008-5472.CAN-09-4487. PMID 20501838.
- 1 2 3 4 Zou, W.; Song, Z.; Guo, Q.; Liu, C.; Zhang, Z.; Zhang, Y. (2011). "Intrathecal Lentiviral-Mediated RNA Interference Targeting PKCγ Attenuates Chronic Constriction Injury–Induced Neuropathic Pain in Rats". Human Gene Therapy 22 (4): 465–475. doi:10.1089/hum.2010.207. PMID 21087146.
- 1 2 Senzer, N.; Barve, M.; Kuhn, J.; Melnyk, A.; Beitsch, P.; Lazar, M.; Lifshitz, S.; Magee, M.; Oh, J.; Mill, S. W.; Bedell, C.; Higgs, C.; Kumar, P.; Yu, Y.; Norvell, F.; Phalon, C.; Taquet, N.; Rao, D. D.; Wang, Z.; Jay, C. M.; Pappen, B. O.; Wallraven, G.; Brunicardi, F. C.; Shanahan, D. M.; Maples, P. B.; Nemunaitis, J. (2011). "Phase I Trial of "bi-shRNAifurin/GMCSF DNA/Autologous Tumor Cell" Vaccine (FANG) in Advanced Cancer". Molecular Therapy 20 (3): 679–686. doi:10.1038/mt.2011.269. PMC 3293620. PMID 22186789.
- 1 2 Millington-Ward, S.; Chadderton, N.; O'Reilly, M.; Palfi, A.; Goldmann, T.; Kilty, C.; Humphries, M.; Wolfrum, U.; Bennett, J.; Humphries, P.; Kenna, P. F.; Farrar, G. J. (2011). "Suppression and Replacement Gene Therapy for Autosomal Dominant Disease in a Murine Model of Dominant Retinitis Pigmentosa". Molecular Therapy 19 (4): 642–649. doi:10.1038/mt.2010.293. PMC 3070095. PMID 21224835.
- ↑ Gu, S.; Jin, L.; Zhang, Y.; Huang, Y.; Zhang, F.; Valdmanis, P. N.; Kay, M. A. (2012). "The Loop Position of shRNAs and Pre-miRNAs is Critical for the Accuracy of Dicer Processing In Vivo". Cell 151 (4): 900–911. doi:10.1016/j.cell.2012.09.042. PMC 3499986. PMID 23141545.
- 1 2 Ringpis, G. E. E.; Shimizu, S.; Arokium, H.; Camba-Colón, J.; Carroll, M. V.; Cortado, R.; Xie, Y.; Kim, P. Y.; Sahakyan, A.; Lowe, E. L.; Narukawa, M.; Kandarian, F. N.; Burke, B. P.; Symonds, G. P.; An, D. S.; Chen, I. S.; Kamata, M. (2012). Speck, Roberto F, ed. "Engineering HIV-1-Resistant T-Cells from Short-Hairpin RNA-Expressing Hematopoietic Stem/Progenitor Cells in Humanized BLT Mice". PLoS ONE 7 (12): e53492. doi:10.1371/journal.pone.0053492. PMC 3534037. PMID 23300932.
- 1 2 "Benitec Biopharma Ltd Annual Report 2012" (PDF). Benitec Biopharma. Retrieved 29 April 2013.
- 1 2 "Proactive Investors Press Release. Benitec Biopharma acquires US-based Tacere Therapeutics. 30 October 2012". Proactive Investors. Retrieved 29 April 2013.
- ↑ "Evaluate Press Release. Genable Technologies executes license agreement with Benitec Biopharma for a genetic eye disease retinitis pigmentosa. 10 July 2012". Evaluate Group. Retrieved 29 April 2013.
- 1 2 "GenomeWeb Press Release. Benitec licenses expressed RNAi tech to Calimmune. 8 March 2012". GenomeWeb. Retrieved 29 April 2013.
- ↑ "Genetic Engineering & Biotechnology News Press Release. UniQure and Benitec enter cross-licensing agreement. 5 December 2012". GEN. Retrieved 29 April 2013.
- 1 2 3 4 5 Benitec Biopharma Ausbiotech presentation March 2012
- ↑ Lavender, H.; Brady, K.; Burden, F.; Delpuech-Adams, O.; Denise, H.; Palmer, A.; Perkins, H.; Savic, B.; Scott, S.; Smith-Burchnell, C.; Troke, P.; Wright, J. F.; Suhy, D.; Corbau, R. (2011). "In Vitro Characterization of the Activity of PF-05095808, a Novel Biological Agent for Hepatitis C Virus Therapy". Antimicrobial Agents and Chemotherapy 56 (3): 1364–1375. doi:10.1128/AAC.05357-11. PMC 3294929. PMID 22203606.
- ↑ Trollet, C.; Anvar, S. Y.; Venema, A.; Hargreaves, I. P.; Foster, K.; Vignaud, A.; Ferry, A.; Negroni, E.; Hourde, C.; Baraibar, M. A.; 't Hoen, P. A. C.; Davies, J. E.; Rubinsztein, D. C.; Heales, S. J.; Mouly, V.; Van Der Maarel, S. M.; Butler-Browne, G.; Raz, V.; Dickson, G. (2010). "Molecular and phenotypic characterization of a mouse model of oculopharyngeal muscular dystrophy reveals severe muscular atrophy restricted to fast glycolytic fibres". Human Molecular Genetics 19 (11): 2191–2207. doi:10.1093/hmg/ddq098. PMID 20207626.
- ↑ Brake, O. T.; Hooft, K. 'T.; Liu, Y. P.; Centlivre, M.; Jasmijn Von Eije, K.; Berkhout, B. (2008). "Lentiviral Vector Design for Multiple shRNA Expression and Durable HIV-1 Inhibition". Molecular Therapy 16 (3): 557–564. doi:10.1038/sj.mt.6300382. PMID 18180777.
- ↑ "US NIH ClincalTrials.gov. Safety study of a dual anti-HIV gene transfer construct to treat HIV-1 infection". ClinicalTrials.gov. Retrieved 29 April 2013.
- ↑ Woods, N. B.; Bottero, V.; Schmidt, M.; Von Kalle, C.; Verma, I. M. (2006). "Gene therapy: Therapeutic gene causing lymphoma". Nature 440 (7088): 1123. doi:10.1038/4401123a. PMID 16641981.
- ↑ Wang, G. P.; Levine, B. L.; Binder, G. K.; Berry, C. C.; Malani, N.; McGarrity, G.; Tebas, P.; June, C. H.; Bushman, F. D. (2009). "Analysis of Lentiviral Vector Integration in HIV+ Study Subjects Receiving Autologous Infusions of Gene Modified CD4+ T Cells". Molecular Therapy 17 (5): 844–850. doi:10.1038/mt.2009.16. PMC 2835137. PMID 19259065.
- ↑ Kleinman, M. E.; Yamada, K.; Takeda, A.; Chandrasekaran, V.; Nozaki, M.; Baffi, J. Z.; Albuquerque, R. J. C.; Yamasaki, S.; Itaya, M.; Pan, Y.; Appukuttan, B.; Gibbs, D.; Yang, Z.; Karikó, K.; Ambati, B. K.; Wilgus, T. A.; Dipietro, L. A.; Sakurai, E.; Zhang, K.; Smith, J. R.; Taylor, E. W.; Ambati, J. (2008). "Sequence- and target-independent angiogenesis suppression by siRNA via TLR3". Nature 452 (7187): 591–597. doi:10.1038/nature06765. PMC 2642938. PMID 18368052.
- ↑ McBride, J. L.; Boudreau, R. L.; Harper, S. Q.; Staber, P. D.; Monteys, A. M.; Martins, I.; Gilmore, B. L.; Burstein, H.; Peluso, R. W.; Polisky, B.; Carter, B. J.; Davidson, B. L. (2008). "Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: Implications for the therapeutic development of RNAi". Proceedings of the National Academy of Sciences 105 (15): 5868–5873. doi:10.1073/pnas.0801775105. PMC 2311380. PMID 18398004.
- ↑ Jackson, A. L.; Bartz, S. R.; Schelter, J.; Kobayashi, S. V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P. S. (2003). "Expression profiling reveals off-target gene regulation by RNAi". Nature Biotechnology 21 (6): 635–637. doi:10.1038/nbt831. PMID 12754523.
External links
- Benitec Biopharma, Balmain, NSW, Australia
- Biomic Biotechnologies, Nantong, China
- Calimmune, Tucson, AZ, US
- Center for Infection and Immunity Amsterdam (CINIMA), University of Amsterdam, The Netherlands
- Children’s Cancer Institute Australia, University of New South Wales, NSW, Australia
- City of Hope National Medical Center, California, CA, US
- CSIRO, VIC, Australia
- Genable Technologies, Dublin, Ireland
- The Institut de Myologie, Paris, France
- The Royal Holloway, London, UK
- Stanford University, Stanford, CA, US
- Tacere Therapeutics Inc, San Francisco, CA, US
- UniQure, Amsterdam, The Netherlands
- University of Queensland, QLD, Australia