DNA laddering
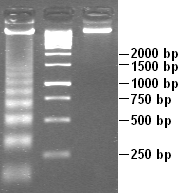
DNA laddering is a feature that can be observed when DNA fragments, resulting from apoptotic DNA fragmentation, are visualised after separation by gel electrophoresis. It was first described in 1980 by Andrew Wyllie at the University of Edinburgh Medical School.[1]
DNA laddering is a distinctive feature of DNA degraded by caspase-activated DNase (CAD), which is a key event during apoptosis. CAD cleaves genomic DNA at internucleosomal linker regions, resulting in DNA fragments that are multiples of 180–185 base-pairs in length. Separation of the fragments by agarose gel electrophoresis and subsequent visualisation, for example by ethidium bromide staining, results in a characteristic "ladder" pattern.
While most of the morphological features of apoptotic cells are short-lived, DNA laddering can be used as final state read-out method and has therefore become a reliable method to distinguish apoptosis from necrosis.[2]
References
- ↑ Wyllie AH (1980-04-10). "Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation". Nature 284 (5756): 555–556. doi:10.1038/284555a0. ISSN 0028-0836. PMID 6245367.
- ↑ Iwata M, Myerson D, Torok-Storb B, Zager RA. (December 1994). "An evaluation of renal tubular DNA laddering in response to oxygen deprivation and oxidant injury". Journal of the American Society of Nephrology 5 (6): 1307–1313. ISSN 1046-6673. PMID 7893995.
Further reading
Yeung, M. C. (2002). "Accelerated apoptotic DNA laddering protocol". BioTechniques 33 (4): 734, 736. PMID 12398177.