Cytomegalovirus
| Cytomegalovirus | |
|---|---|
| Classification and external resources | |
| Specialty | Infectious disease |
| ICD-10 | B25 |
| ICD-9-CM | 078.5 |
| MedlinePlus | 000568 |
| Patient UK | Cytomegalovirus |
| MeSH | D003586 |
| Cytomegalovirus | |
|---|---|
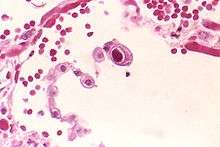 | |
| Typical "owl eye" inclusion indicating CMV infection of a lung pneumocyte[1] | |
| Virus classification | |
| Group: | Group I (dsDNA) |
| Order: | Herpesvirales |
| Family: | Herpesviridae |
| Subfamily: | Betaherpesvirinae |
| Genus: | Cytomegalovirus |
| Type species | |
| Human cytomegalovirus | |
Cytomegalovirus (from the Greek cyto-, "cell", and megalo-, "large") is a genus of viruses in the order Herpesvirales, in the family Herpesviridae, in the subfamily Betaherpesvirinae. Human and monkeys serve as natural hosts. There are currently eight species in this genus including the type species human herpesvirus 5. Diseases associated with HHV-5 include mononucleosis, and pneumonias.[2][3] It is typically abbreviated as CMV.
The species that infects humans is commonly known as human CMV (HCMV) or human herpesvirus-5 (HHV-5), and is the most studied of all cytomegaloviruses.[4] Within Herpesviridae, CMV belongs to the Betaherpesvirinae subfamily, which also includes the genera Muromegalovirus and Roseolovirus (HHV-6 and HHV-7).[5] It is related to other herpesviruses within the subfamilies of Alphaherpesvirinae that includes herpes simplex viruses (HSV)-1 and -2 and varicella-zoster virus (VZV), and the Gammaherpesvirinae subfamily that includes Epstein–Barr virus.[4]
All herpesviruses share a characteristic ability to remain latent within the body over long periods. Although they may be found throughout the body, CMV infections are frequently associated with the salivary glands in humans and other mammals.[5] Other CMV viruses are found in several mammal species, but species isolated from animals differ from HCMV in terms of genomic structure, and have not been reported to cause human disease.
Taxonomy
Group: dsDNA
- Family: Herpesviridae
- Sub-Family: Betaherpesvirinae
- Genus: Cytomegalovirus
- Aotine herpesvirus 1
- Cebine herpesvirus 1
- Cercopithecine herpesvirus 5
- Human herpesvirus 5
- Macacine herpesvirus 3
- Panine herpesvirus 2
- Papiine herpesvirus 3
- Saimiriine herpesvirus 4
Species
| Scientific Name | Host | Common Name |
|---|---|---|
|
Human herpesvirus 5 (HHV-5)
|
Human
|
Human CMV (HCMV)
|
Several species of Cytomegalovirus have been identified and classified for different mammals.[5] The most studied is Human cytomegalovirus (HCMV), which is also known as Human herpesvirus 5 (HHV-5). Other primate CMV species include Chimpanzee cytomegalovirus (CCMV) that infects chimpanzees and orangutans, and Simian cytomegalovirus (SCCMV) and Rhesus cytomegalovirus (RhCMV) that infect macaques; CCMV is known as both Panine herpesvirus 2 (PaHV-2) and Pongine herpesvirus-4 (PoHV-4). SCCMV is called Cercopithecine herpesvirus-5 (CeHV-5) and RhCMV, Cercopithecine herpesvirus 8 (CeHV-8). A further two viruses found in the night monkey are tentatively placed in the Cytomegalovirus genus, and are called Herpesvirus aotus 1 and Herpesvirus aotus 3. Rodents also have viruses previously called cytomegaloviruses that are now reclassified under the genus Muromegalovirus; this genus contains Mouse cytomegalovirus (MCMV) is also known as Murid herpesvirus 1 (MuHV-1) and the closely related Murid herpesvirus 2 (MuHV-2) that is found in rats. In addition, there many other viral species with the name Cytomegalovirus identified in distinct mammals that are as yet not completely classified; these were predominantly isolated from primates and rodents.
Structure
Viruses in Cytomegalovirus are enveloped, with icosahedral, Spherical to pleomorphic, and Round geometries, and T=16 symmetry. The diameter is around 150-200 nm. Genomes are linear and non-segmented, around 200kb in length.[2]
| Genus | Structure | Symmetry | Capsid | Genomic Arrangement | Genomic Segmentation |
|---|---|---|---|---|---|
| Cytomegalovirus | Spherical Pleomorphic | T=16 | Enveloped | Linear | Monopartite |
Life Cycle
Viral replication is nuclear, and is lysogenic. Entry into the host cell is achieved by attachment of the viral glycoproteins to host receptors, which mediates endocytosis. Replication follows the dsDNA bidirectional replication model. DNA templated transcription, with some alternative splicing mechanism is the method of transcription. Translation takes place by leaky scanning. The virus exits the host cell by nuclear egress, and budding. Human and monkeys serve as the natural host. Transmission routes are contact, urine, and saliva.[2]
| Genus | Host Details | Tissue Tropism | Entry Details | Release Details | Replication Site | Assembly Site | Transmission |
|---|---|---|---|---|---|---|---|
| Cytomegalovirus | Humans; monkeys | Epithelial mucosa | Glycoprotiens | Budding | Nucleus | Nucleus | Urine; saliva |
Genetic engineering
The CMV promoter is commonly included in vectors used in genetic engineering work conducted in mammalian cells, as it is a strong promoter and drives constitutive expression of genes under its control.[6]
References
- ↑ Mattes FM, McLaughlin JE, Emery VC, Clark DA, Griffiths PD (August 2000). "Histopathological detection of owl's eye inclusions is still specific for cytomegalovirus in the era of human herpesviruses 6 and 7". J. Clin. Pathol. 53 (8): 612–4. doi:10.1136/jcp.53.8.612. PMC 1762915. PMID 11002765.
- 1 2 3 "Viral Zone". ExPASy. Retrieved 15 June 2015.
- 1 2 ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- 1 2 Ryan KJ, Ray CG (editors) (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 556; 566–9. ISBN 0-8385-8529-9.
- 1 2 3 Koichi Yamanishi; Arvin, Ann M.; Gabriella Campadelli-Fiume; Edward Mocarski; Moore, Patrick; Roizman, Bernard; Whitley, Richard (2007). Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge, UK: Cambridge University Press. ISBN 0-521-82714-0.
- ↑ Kendall Morgan for Addgene Blog. Apr 3, 2014 Plasmids 101: The Promoter Region – Let's Go!
External links
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||