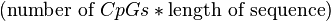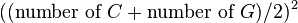CpG site


The CpG sites or CG sites are regions of DNA where a cytosine nucleotide is followed by a guanine nucleotide in the linear sequence of bases along its 5' → 3' direction. CpG is shorthand for 5'—C—phosphate—G—3' , that is, cytosine and guanine separated by only one phosphate; phosphate links any two nucleosides together in DNA. The CpG notation is used to distinguish this single-stranded linear sequence from the CG base-pairing of cytosine and guanine for double-stranded sequences. The CpG notation is therefore to be interpreted as the cytosine being 5 prime to the guanine base. CpG should not be confused with GpC, the latter meaning that a guanine is followed by a cytosine in the 5' → 3' direction of a single-stranded sequence.
Cytosines in CpG dinucleotides can be methylated to form 5-methylcytosine. In mammals, methylating the cytosine within a gene can change its expression, a mechanism that is part of a larger field of science studying gene regulation that is called epigenetics. Enzymes that add a methyl group are called DNA methyltransferases.
In mammals, 70% to 80% of CpG cytosines are methylated.[1]
Unmethylated CpG dinucleotide sites can be detected by Toll-like receptor 9[2] (TLR 9) on plasmacytoid dendritic cells, monocytes, natural killer (NK) cells, and B cells in humans. This is used to detect intracellular Viral infection.
Frequency in vertebrates
CpG dinucleotides have long been observed to occur with a much lower frequency in the sequence of vertebrate genomes than would be expected due to random chance. For example, in the human genome, which has a 42% GC content, a pair of nucleotides consisting of cytosine followed by guanine would be expected to occur 0.21 * 0.21 = 4.41% of the time. The frequency of CpG dinucleotides in human genomes is 1% — less than one-quarter of the expected frequency. Scarano et al. proposed that the CpG deficiency is due to an increased vulnerability of methylcytosines to spontaneously deaminate to thymine in genomes with CpG cytosine methylation.[3]
CpG islands

CpG islands (or CG islands) are regions with a high frequency of CpG sites, though objective definitions for CpG islands are limited. The usual formal definition of a CpG island is a region with at least 200 bp, and a GC percentage that is greater than 50 %, and with an observed-to-expected CpG ratio that is greater than 60 %. The "observed-to-expected CpG ratio" can be derived where the observed is calculated as:
and the expected as:
or
Many genes in mammalian genomes have CpG islands associated with the start of the gene[6] (promoter regions). Because of this, the presence of a CpG island is used to help in the prediction and annotation of genes.
In mammalian genomes, CpG islands are typically 300-3,000 base pairs in length, and have been found in or near approximately 40% of promoters of mammalian genes.[7] About 70% of human promoters have a high CpG content. Given the frequency of GC two-nucleotide sequences, the number of CpG dinucleotides is much lower than would be expected.[5]
A 2002 study revised the rules of CpG island prediction to exclude other GC-rich genomic sequences such as Alu repeats. Based on an extensive search on the complete sequences of human chromosomes 21 and 22, DNA regions greater than 500 bp were found more likely to be the "true" CpG islands associated with the 5' regions of genes if they had a GC content greater than 55%, and an observed-to-expected CpG ratio of 65%.[8]
CpG islands are characterized by CpG dinucleotide content of at least 60% of that which would be statistically expected (~4–6%), whereas the rest of the genome has much lower CpG frequency (~1%), a phenomenon called CG suppression. Unlike CpG sites in the coding region of a gene, in most instances the CpG sites in the CpG islands of promoters are unmethylated if the genes are expressed. This observation led to the speculation that methylation of CpG sites in the promoter of a gene may inhibit gene expression. Methylation, along with histone modification, is central to imprinting.[9] Most of the methylation differences between tissues, or between normal and cancer samples, occur a short distance from the CpG islands (at "CpG island shores") rather than in the islands themselves.[10]
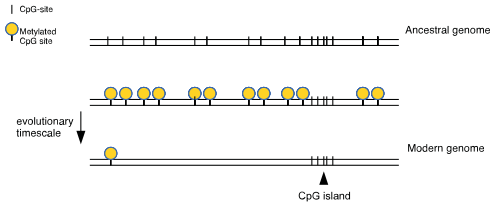
CpG islands typically occur at or near the transcription start site of genes, particularly housekeeping genes, in vertebrates.[5] A C (cytosine) base followed immediately by a G (guanine) base (a CpG) is rare in vertebrate DNA because the cytosines in such an arrangement tend to be methylated. This methylation helps distinguish the newly synthesized DNA strand from the parent strand, which aids in the final stages of DNA proofreading after duplication. However, over time methylated cytosines tend to turn into thymines because of spontaneous deamination. There is a special enzyme in humans (Thymine-DNA glycosylase, or TDG) that specifically replaces T's from T/G mismatches. However, due to the rarity of CpGs, it is theorised to be insufficiently effective in preventing a possibly rapid mutation of the dinucleotides. The existence of CpG islands is usually explained by the existence of selective forces for relatively high CpG content, or low levels of methylation in that genomic area, perhaps having to do with the regulation of gene expression. Recently a study showed that most CpG islands are a result of non-selective forces.[11]
Methylation, silencing, cancer, and aging
Methylation of CpG sites within the promoters of genes can lead to their silencing, a feature found in a number of human cancers (for example the silencing of tumor suppressor genes). In contrast, the hypomethylation of CpG sites has been associated with the over-expression of oncogenes within cancer cells.[12]
Since age has a strong effect on DNA methylation levels on tens of thousands of CpG sites, one can define a highly accurate biological clock (referred to as epigenetic clock or DNA methylation age) in humans and chimpanzees.[13]
See also
- TLR9, detector of unmethylated CpG sites
- DNA methylation age
References
- ↑ Jabbari K, Bernardi G (May 2004). "Cytosine methylation and CpG, TpG (CpA) and TpA frequencies". Gene 333: 143–9. doi:10.1016/j.gene.2004.02.043. PMID 15177689.
- ↑ Ramirez-Ortiz ZG, Specht CA, Wang JP, Lee CK, Bartholomeu DC, Gazzinelli RT, Levitz SM (2008). "Toll-like receptor 9-dependent immune activation by unmethylated CpG motifs in Aspergillus fumigatus DNA". Infect Immun. 76 (5): 2123–2129. doi:10.1128/IAI.00047-08. PMC 2346696. PMID 18332208.
- ↑ Scarano E, Iaccarino M, Grippo P, Parisi E (1967). "The heterogeneity of thymine methyl group origin in DNA pyrimidine isostichs of developing sea urchin embryos". Proc. Natl. Acad. Sci. USA 57 (5): 1394–400. doi:10.1073/pnas.57.5.1394. PMC 224485. PMID 5231746.
- ↑ Gardiner-Garden M, Frommer M (1987). "CpG islands in vertebrate genomes". Journal of Molecular Biology 196 (2): 261–282. doi:10.1016/0022-2836(87)90689-9. PMID 3656447.
- 1 2 3 Saxonov S, Berg P, Brutlag DL (2006). "A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters". Proc Natl Acad Sci USA 103 (5): 1412–1417. doi:10.1073/pnas.0510310103. PMC 1345710. PMID 16432200.
- ↑ Hartl DL, Jones EW (2005). Genetics: Analysis of Genes and Genomes (6 ed.). Missisauga: Jones & Bartlett, Canada. p. 477. ISBN 0-7637-1511-5.
- ↑ Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA (2005). "Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level". Nucleic Acids Res 33 (20): e176. doi:10.1093/nar/gni180. PMC 1292996. PMID 16314307.
- ↑ Takai D, Jones PA (2002). "Comprehensive analysis of CpG islands in human chromosomes 21 and 22.". Proc Natl Acad Sci USA 99 (6): 3740–5. doi:10.1073/pnas.052410099. PMC 122594. PMID 11891299.
- ↑ Feil R, Berger F (2007). "Convergent evolution of genomic imprinting in plants and mammals". Trends Genet 23 (4): 192–199. doi:10.1016/j.tig.2007.02.004. PMID 17316885.
- ↑ Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP (2009). "The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores". Nature Genetics 41 (2): 178–186. doi:10.1038/ng.298. PMC 2729128. PMID 19151715.
- ↑ Cohen N, Kenigsberg E, Tanay A (2011). "Primate CpG Islands Are Maintained by Heterogeneous Evolutionary Regimes Involving Minimal Selection". Cell 145 (5): 773–786. doi:10.1016/j.cell.2011.04.024. PMID 21620139.
- ↑ Jones PA, Laird PW (February 1999). "Cancer epigenetics comes of age". Nat. Genet. 21 (2): 163–7. doi:10.1038/5947. PMID 9988266.
- ↑ Horvath S (2013). "DNA methylation age of human tissues and cell types". Genome Biology 14 (10): R115. doi:10.1186/gb-2013-14-10-r115. PMC 4015143. PMID 24138928.