Biosensor
A biosensor is an analytical device, used for the detection of an analyte, that combines a biological component with a physicochemical detector.[1][2]
- the sensitive biological element (e.g. tissue, microorganisms, organelles, cell receptors, enzymes, antibodies, nucleic acids, etc.), a biologically derived material or biomimetic component that interacts (binds or recognizes) the analyte under study. The biologically sensitive elements can also be created by biological engineering.
- the transducer or the detector element (works in a physicochemical way; optical, piezoelectric, electrochemical, etc.) that transforms the signal resulting from the interaction of the analyte with the biological element into another signal (i.e., transduces) that can be more easily measured and quantified;
- biosensor reader device with the associated electronics or signal processors that are primarily responsible for the display of the results in a user-friendly way.[3] This sometimes accounts for the most expensive part of the sensor device, however it is possible to generate a user friendly display that includes transducer and sensitive element(see Holographic Sensor). The readers are usually custom-designed and manufactured to suit the different working principles of biosensors. Known manufacturers of biosensor electronic readers include PalmSens, Gwent Biotechnology Systems and Rapid Labs.
Examples and applications
A common example of a commercial biosensor is the blood glucose biosensor, which uses the enzyme glucose oxidase to break blood glucose down. In doing so it first oxidizes glucose and uses two electrons to reduce the FAD (a component of the enzyme) to FADH2. This in turn is oxidized by the electrode in a number of steps. The resulting current is a measure of the concentration of glucose. In this case, the electrode is the transducer and the enzyme is the biologically active component.
Recently, arrays of many different detector molecules have been applied in so called electronic nose devices, where the pattern of response from the detectors is used to fingerprint a substance.[4] In the Wasp Hound odor-detector, the mechanical element is a video camera and the biological element is five parasitic wasps who have been conditioned to swarm in response to the presence of a specific chemical.[5] Current commercial electronic noses, however, do not use biological elements.
A canary in a cage, as used by miners to warn of gas, could be considered a biosensor. Many of today's biosensor applications are similar, in that they use organisms which respond to toxic substances at a much lower concentrations than humans can detect to warn of their presence. Such devices can be used in environmental monitoring,[6] trace gas detection and in water treatment facilities.
Many optical biosensors are based on the phenomenon of surface plasmon resonance (SPR) techniques.[7][8] This utilises a property of and other materials; specifically that a thin layer of gold on a high refractive index glass surface can absorb laser light, producing electron waves (surface plasmons) on the gold surface. This occurs only at a specific angle and wavelength of incident light and is highly dependent on the surface of the gold, such that binding of a target analyte to a receptor on the gold surface produces a measurable signal.
Surface plasmon resonance sensors operate using a sensor chip consisting of a plastic cassette supporting a glass plate, one side of which is coated with a microscopic layer of gold. This side contacts the optical detection apparatus of the instrument. The opposite side is then contacted with a microfluidic flow system. The contact with the flow system creates channels across which reagents can be passed in solution. This side of the glass sensor chip can be modified in a number of ways, to allow easy attachment of molecules of interest. Normally it is coated in carboxymethyl dextran or similar compound.
Light of a fixed wavelength is reflected off the gold side of the chip at the angle of total internal reflection, and detected inside the instrument. The angle of incident light is varied in order to match the evanescent wave propagation rate with the propagation rate of the surface plasmon plaritons.[9] This induces the evanescent wave to penetrate through the glass plate and some distance into the liquid flowing over the surface.
The refractive index at the flow side of the chip surface has a direct influence on the behaviour of the light reflected off the gold side. Binding to the flow side of the chip has an effect on the refractive index and in this way biological interactions can be measured to a high degree of sensitivity with some sort of energy. The refractive index of the medium near the surface changes when biomolecules attach to the surface, and the SPR angle varies as a function of this change.
Other evanescent wave biosensors have been commercialised using waveguides where the propagation constant through the waveguide is changed by the absorption of molecules to the waveguide surface. One such example, Dual Polarisation Interferometry uses a buried waveguide as a reference against which the change in propagation constant is measured. Other configurations such as the Mach-Zehnder have reference arms lithographically defined on a substrate. Higher levels of integration can be achieved using resonator geometries where the resonant frequency of a ring resonator changes when molecules are absorbed.[10][11]
Other optical biosensors are mainly based on changes in absorbance or fluorescence of an appropriate indicator compound and do not need a total internal reflection geometry. For example, a fully operational prototype device detecting casein in milk has been fabricated. The device is based on detecting changes in absorption of a gold layer.[12] A widely used research tool, the micro-array, can also be considered a biosensor.
Nanobiosensors use an immobilized bioreceptor probe that is selective for target analyte molecules. Nanomaterials are exquisitely sensitive chemical and biological sensors. Nanoscale materials demonstrate unique properties. Their large surface area to volume ratio can achieve rapid and low cost reactions, using a variety of designs.[13]
Biological biosensors often incorporate a genetically modified form of a native protein or enzyme. The protein is configured to detect a specific analyte and the ensuing signal is read by a detection instrument such as a fluorometer or luminometer. An example of a recently developed biosensor is one for detecting cytosolic concentration of the analyte cAMP (cyclic adenosine monophosphate), a second messenger involved in cellular signaling triggered by ligands interacting with receptors on the cell membrane.[14] Similar systems have been created to study cellular responses to native ligands or xenobiotics (toxins or small molecule inhibitors). Such "assays" are commonly used in drug discovery development by pharmaceutical and biotechnology companies. Most cAMP assays in current use require lysis of the cells prior to measurement of cAMP. A live-cell biosensor for cAMP can be used in non-lysed cells with the additional advantage of multiple reads to study the kinetics of receptor response.
The biosensor system
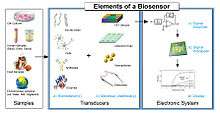
A biosensor typically consists of a bio-recognition component, biotransducer component, and electronic system which include a signal amplifier, processor, and display. Transducers and electronics can be combined, e.g., in CMOS-based microsensor systems.[15][16] The recognition component, often called a bioreceptor, uses biomolecules from organisms or receptors modeled after biological systems to interact with the analyte of interest. This interaction is measured by the biotransducer which outputs a measurable signal proportional to the presence of the target analyte in the sample. The general aim of the design of a biosensor is to enable quick, convenient testing at the point of concern or care where the sample was procured.[17]
Bioreceptors
In a biosensor, the bioreceptor is designed to interact with the specific analyte of interest to produce an effect measurable by the transducer. High selectivity for the analyte among a matrix of other chemical or biological components is a key requirement of the bioreceptor. While the type of biomolecule used can vary widely, biosensors can be classified according to common types bioreceptor interactions involving: anitbody/antigen, enzymes, nucleic acids/DNA, cellular structures/cells, or biomimetic materials.[18]
Antibody/antigen interactions
An immunosensor utilizes the very specific binding affinity of antibodies for a specific compound or antigen. The specific nature of the antibody antigen interaction is analogous to a lock and key fit in that the antigen will only bind to the antibody if it has the correct conformation. Binding events result in a physicochemical change that in combination with a tracer, such as a fluorescent molecules, enzymes, or radioisotopes, can generate a signal. There are limitations with using antibodies in sensors: 1.The antibody binding capacity is strongly dependent on assay conditions (e.g. pH and temperature) and 2. The antibody-antigen interaction is generally irreversible. However, it has been shown that binding can be disrupted by chaotropic reagents, organic solvents, or even ultrasonic radiation.[19]
Enzymatic interactions
The specific binding capabilities and catalytic activity of enzymes make them popular bioreceptors. Analyte recognition is enabled through several possible mechanisms: 1) the enzyme converting the analyte into a product that is sensor-detectable, 2) detecting enzyme inhibition or activation by the analyte,[20] or 3) monitoring modification of enzyme properties resulting from interaction with the analyte.[19] The main reasons for the common use of enzymes in biosensors are: 1) ability to catalyze a large number of reactions; 2) potential to detect a group of analytes (substrates, products, inhibitors, and modulators of the catalytic activity); and 3) suitability with several different transduction methods for detecting the analyte. Notably, since enzymes are not consumed in reactions, the biosensor can easily be used continuously. The catalytic activity of enzymes also allows lower limits of detection compared to common binding techniques. However, the sensor's lifetime is limited by the stability of the enzyme.
Nucleic acid interactions
Biosensors that employ nucleic acid interactions can be referred to as genosensors. The recognition process is based on the principle of complementary base pairing, adenine:thymine and cytosine:guanine in DNA. If the target nucleic acid sequence is known,complementary sequences can be synthesized, labeled, and then immobilized on the sensor. The hybridization probes can then base pair with the target sequences, generating an optical signal. The favored transduction principle employed in this type of sensor has been optical detection.[19]
Epigenetics
It has been proposed that properly optimized integrated optical resonators can be exploited for detecting epigenetic modifications (e.g. DNA methylation, histone post-translational modifications) in body fluids from patients affected by cancer or other diseases.[21] Photonic biosensors with ultra-sensitivity are nowadays being developed at a research level to easily detect cancerous cells within the patient's urine.[22]
Organelles
Organelles form separate compartments inside cells and usually perform function independently. Different kinds of organelles have various metabolic pathways and contain enzymes to fulfill its function. Commonly used organelles include lysosome, chloroplast and mitochondria. The spatial-temporal distribution pattern of calcium is closed related to ubiquitous signaling pathway. Mitochondria actively participate in the metabolism of calcium ions to control the function and also modulate the calcium related signaling pathways. Experiments have proved that mitochondria have the ability to respond to high calcium concentration generated in the proximity by opening the calcium channel.[23] In this way, mitochondria can be used to detect the calcium concentration in medium and the detection is very sensitive due to high spatial resolution. Another application of mitochondria is used for detection of water pollution. Detergent compounds’ toxicity will damage the cell and subcellular structure including mitochondria. The detergents will cause a swelling effect which could be measured by an absorbance change. Experiment data shows the change rate is proportional to the detergent concentration, providing a high standard for detection accuracy.[24]
Cells
Cells are often used in bioreceptors because they are sensitive to surrounding environment and they can respond to all kinds of stimulants. Cells tend to attach to the surface so they can be easily immobilized. Compared to organelles they remain active for longer period and the reproducibility makes them reusable. They are commonly used to detect global parameter like stress condition, toxicity and organic derivatives. They can also be used to monitor the treatment effect of drugs. One application is to use cells to determine herbicides which are main aquatic contaminant.[25] Microalgae are entrapped on a quartz microfiber and the chlorophyll fluorescence modified by herbicides is collected at the tip of an optical fiber bundle and transmitted to a fluorimeter. The algae are continuously cultured to get optimized measurement. Results show that detection limit of certain herbicide can reach sub-ppb concentration level. Some cells can also be used to monitor the microbial corrosion.[26] Pseudomonas sp. is isolated form corroded material surface and immobilized on acetylcellulose membrane. The respiration activity is determined by measuring oxygen consumption. There is linear relationship between the current generated and the concentration of sulfuric acid. The response time is related to the loading of cells and surrounding environments and can be controlled to no more than 5min.
Tissue
Tissues are used for biosensor for the abundance of enzymes existed. Advantages of tissues as biosensors include the following:[27] 1)easier to immobilize compared to cells and organelles 2)the higher activity and stability from maintain enzymes in natural environment 3)the availability and low price 4)the avoidance of tedious work of extraction, centrifuge and purification of enzymes 5)necessary cofactors for enzyme to function exists 6)the diversity providing a wide range of choice concerning different objectives. There also exists some disadvantages of tissues like the lack of specificity due to the interference of other enzymes and longer response time due to transport barrier.
Surface attachment of the biological elements
An important part in a biosensor is to attach the biological elements (small molecules/protein/cells) to the surface of the sensor (be it metal, polymer or glass). The simplest way is to functionalize the surface in order to coat it with the biological elements. This can be done by polylysine, aminosilane, epoxysilane or nitrocellulose in the case of silicon chips/silica glass. Subsequently the bound biological agent may be for example fixed by Layer by layer depositation of alternatively charged polymer coatings[28]
Alternatively three-dimensional lattices (hydrogel/xerogel) can be used to chemically or physically entrap these (where by chemically entraped it is meant that the biological element is kept in place by a strong bond, while physically they are kept in place being unable to pass through the pores of the gel matrix). The most commonly used hydrogel is sol-gel, a glassy silica generated by polymerization of silicate monomers (added as tetra alkyl orthosilicates, such as TMOS or TEOS) in the presence of the biological elements (along with other stabilizing polymers, such as PEG) in the case of physical entrapment.[29]
Another group of hydrogels, which set under conditions suitable for cells or protein, are acrylate hydrogel, which polymerize upon radical initiation. One type of radical initiator is a peroxide radical, typically generated by combining a persulfate with TEMED (Polyacrylamide gel are also commonly used for protein electrophoresis),[30] alternatively light can be used in combination with a photoinitiator, such as DMPA (2,2-dimethoxy-2-phenylacetophenone).[31] Smart materials that mimic the biological components of a sensor can also be classified as biosensors using only the active or catalytic site or analogous configurations of a biomolecule.[32]
Biotransducer
Biosensors can be classified by their biotransducer type. The most common types of biotransducers used in biosensors are 1) electrochemical biosensors, 2) optical biosensors, 3)electronic biosensors, 4)piezoelectric biosensors, 5) gravimetric biosensors, 6) pyroelectric biosensors.

Electrochemical
Electrochemical biosensors are normally based on enzymatic catalysis of a reaction that produces or consumes electrons (such enzymes are rightly called redox enzymes). The sensor substrate usually contains three electrodes; a reference electrode, a working electrode and a counter electrode. The target analyte is involved in the reaction that takes place on the active electrode surface, and the reaction may cause either electron transfer across the double layer (producing a current) or can contribute to the double layer potential (producing a voltage). We can either measure the current (rate of flow of electrons is now proportional to the analyte concentration) at a fixed potential or the potential can be measured at zero current (this gives a logarithmic response). Note that potential of the working or active electrode is space charge sensitive and this is often used. Further, the label-free and direct electrical detection of small peptides and proteins is possible by their intrinsic charges using biofunctionalized ion-sensitive field-effect transistors.[33]
Another example, the potentiometric biosensor, (potential produced at zero current) gives a logarithmic response with a high dynamic range. Such biosensors are often made by screen printing the electrode patterns on a plastic substrate, coated with a conducting polymer and then some protein (enzyme or antibody) is attached. They have only two electrodes and are extremely sensitive and robust. They enable the detection of analytes at levels previously only achievable by HPLC and LC/MS and without rigorous sample preparation. All biosensors usually involve minimal sample preparation as the biological sensing component is highly selective for the analyte concerned. The signal is produced by electrochemical and physical changes in the conducting polymer layer due to changes occurring at the surface of the sensor. Such changes can be attributed to ionic strength, pH, hydration and redox reactions, the latter due to the enzyme label turning over a substrate (). Field effect transistors, in which the gate region has been modified with an enzyme or antibody, can also detect very low concentrations of various analytes as the binding of the analyte to the gate region of the FET cause a change in the drain-source current.
Ion channel switch


The use of ion channels has been shown to offer highly sensitive detection
of target biological molecules.[34] By embedding the ion channels in supported or tethered bilayer membranes (t-BLM) attached to a gold electrode, an electrical circuit is created. Capture molecules such as antibodies can be bound to the ion channel so that the binding of the target molecule controls the ion flow through the channel. This results in a measurable change in the electrical conduction which is proportional to the concentration of the target.
An Ion Channel Switch (ICS) biosensor can be created using gramicidin, a dimeric peptide channel, in a tethered bilayer membrane.[35] One peptide of gramicidin, with attached antibody, is mobile and one is fixed. Breaking the dimer stops the ionic current through the membrane. The magnitude of the change in electrical signal is greatly increased by separating the membrane from the metal surface using a hydrophilic spacer.
Quantitative detection of an extensive class of target species, including proteins, bacteria, drug and toxins has been demonstrated using different membrane and capture configurations.[36][37]
Others
Piezoelectric sensors utilise crystals which undergo an elastic deformation when an electrical potential is applied to them. An alternating potential (A.C.) produces a standing wave in the crystal at a characteristic frequency. This frequency is highly dependent on the elastic properties of the crystal, such that if a crystal is coated with a biological recognition element the binding of a (large) target analyte to a receptor will produce a change in the resonance frequency, which gives a binding signal. In a mode that uses surface acoustic waves (SAW), the sensitivity is greatly increased. This is a specialised application of the Quartz crystal microbalance as a biosensor
Thermometric and magnetic based biosensors are rare.
Placement of biosensors
An in-vivo biosensor is one that functions inside the body. Biocompatibility concerns follow with the creation of an in-vivo biosensor. That is, an initial inflammatory response occurring after the implantation. The second concern is the long-term interaction with the body during the intended period of the device’s use.[38] Another issue that arises is failure. If there is failure, the device must be removed and replaced, causing additional surgery. An example for application of an in-vivo biosensor is insulin monitoring within the body.
An in-vitro biosensor is a sensor that takes place in a test tube, culture dish, or elsewhere outside a living organism. The sensor uses a biological element, such as an enzyme capable of recognizing or signaling a biochemical change in solution. A transducer is then used to convert the biochemical signal to a quantifiable signal. An example of an in-vitro biosensor is an enzyme-conductimetric biosensor for glucose monitoring.
An at-line biosensor is used in a production line where a sample can be taken, tested, and a decision can be made whether or not the continuation of the production should occur. An example of an at-line biosensor is the monitoring of lactose in a dairy processing plant.[39]
The in line biosensor can be placed within a production line to monitor a variable with continuous production and can be automated. The in-line biosensor becomes another step in the process line. An application of an in-line biosensor is for water purification.
There is a challenge to create a biosensor that can be taken straight to the “point of concern”, that is the location where the test is needed. The elimination of lab testing can save time and money. An application of a point-of-concern biosensor can be for the testing of HIV virus in third world countries where it is difficult for the patients to be tested. A biosensor can be sent directly to the location and a quick and easy test can be used.
Applications
There are many potential applications of biosensors of various types. The main requirements for a biosensor approach to be valuable in terms of research and commercial applications are the identification of a target molecule, availability of a suitable biological recognition element, and the potential for disposable portable detection systems to be preferred to sensitive laboratory-based techniques in some situations. Some examples are given below:
- Glucose monitoring in diabetes patients ←historical market driver
- Other medical health related targets
- Environmental applications e.g. the detection of pesticides and river water contaminants such as heavy metal ions[40]
- Remote sensing of airborne bacteria e.g. in counter-bioterrorist activities
- Remote sensing of water quality in coastal waters by describing online different aspects of clam ethology (biological rhythms, growth rates, spawning or death records) in groups of abandoned bivalves around the world[6]
- Detection of pathogens[41]
- Determining levels of toxic substances before and after bioremediation
- Detection and determining of organophosphate
- Routine analytical measurement of folic acid, biotin, vitamin B12 and pantothenic acid as an alternative to microbiological assay
- Determination of drug residues in food, such as antibiotics and growth promoters, particularly meat and honey.
- Drug discovery and evaluation of biological activity of new compounds.
- Protein engineering in biosensors[42]
- Detection of toxic metabolites such as mycotoxins[43]
Glucose monitoring
Commercially available gluocose monitors rely on amperometric sensing of glucose by means of glucose oxidase, which oxidises glucose producing hydrogen peroxide which is detected by the electrode. To overcome the limitation of amperometric sensors, a flurry of research is present into novel sensing methods, such as fluorescent glucose biosensors.[44]
Interferometric reflectance imaging sensor
The interferometric reflectance imaging sensor (IRIS) is based on the principles of optical interference and consists of a silicon-silicon oxide substrate, standard optics, and low-powered coherent LEDs. When light is illuminated through a low magnification objective onto the layered silicon-silicon oxide substrate, an interferometric signature is produced. As biomass, which has a similar index of refraction as silicon oxide, accumulates on the substrate surface, a change in the interferometric signature occurs and the change can be correlated to a quantifiable mass. Daaboul et al used IRIS to yield a label-free sensitivity of approximately 19 ng/mL.[45] Ahn et al. improved the sensitivity of IRIS through a mass tagging technique.[46]
Since initial publication, IRIS has been adapted to perform various functions. First, IRIS integrated a fluorescence imaging capability into the interferometric imaging instrument as a potential way to address fluorescence protein microarray variability.[47] Briefly, the variation in fluorescence microarrays mainly derives from inconsistent protein immobilization on surfaces and may cause misdiagnoses in allergy microarrays.[48] To correct from any variation in protein immobilization, data acquired in the fluorescence modality is then normalized by the data acquired in the label-free modality.[48] IRIS has also been adapted to perform single nanoparticle counting by simply switching the low magnification objective used for label-free biomass quantification to a higher objective magnification.[49][50] This modality enables size discrimination in complex human biological samples. Monroe et al. used IRIS to quantify protein levels spiked into human whole blood and serum and determined allergen sensitization in characterized human blood samples using zero sample processing.[51] Other practical uses of this device include virus and pathogen detection.[52]
Food analysis
There are several applications of biosensors in food analysis. In the food industry, optics coated with antibodies are commonly used to detect pathogens and food toxins. Commonly, the light system in these biosensors is fluorescence, since this type of optical measurement can greatly amplify the signal.
A range of immuno- and ligand-binding assays for the detection and measurement of small molecules such as water-soluble vitamins and chemical contaminants (drug residues) such as sulfonamides and Beta-agonists have been developed for use on SPR based sensor systems, often adapted from existing ELISA or other immunological assay. These are in widespread use across the food industry.
DNA biosensors
In the future, DNA will find use as a versatile material from which scientists can craft biosensors. DNA biosensors can theoretically be used for medical diagnostics, forensic science, agriculture, or even environmental clean-up efforts. No external monitoring is needed for DNA-based sensing devises. This is a significant advantage. DNA biosensors are complicated mini-machines—consisting of sensing elements, micro lasers, and a signal generator. At the heart of DNA biosensor function is the fact that two strands of DNA stick to each other by virtue of chemical attractive forces. On such a sensor, only an exact fit—that is, two strands that match up at every nucleotide position—gives rise to a fluorescent signal (a glow) that is then transmitted to a signal generator.
Microbial biosensors
Using biological engineering researchers have created many microbial biosensors. An example is the arsenic biosensor. To detect arsenic they use the Ars operon.[53] Using bacteria, researchers can detect pollutants in samples.
Ozone Biosensors
because ozone filters out harmful ultraviolet radiation, the discovery of holes in the ozone layer of the earth's atmosphere has raised concern about how much ultraviolet light reaches the earth's surface. Of particular concern are the questions of how deeply into sea water ultraviolet radiation penetrates and how it affects marine organisms, especially plankton (floating microorganisms) and viruses that attack plankton. Plankton form the base of the marine food chains and are believed to affect our planet's temperature and weather by uptake of CO2 for photosynthesis.
Deneb Karentz, a researcher at the Laboratory of Radio-biology and Environmental Health (University of California in San Francisco) has devised a simple method for measuring ultraviolet penetration and intensity. Working in the Antarctic Ocean, she submerfed to various depths thin plastic bags containing special strains of E. coli that are almost totally unable to repair ultraviolet radiation damage to their DNA. Bacterial death rates in these bags were compared with rates in unexposed control bags of the same organism. The bacterial "biosensors" revealed constant significant ultraviolet damage at depths of 10 m and frequently at 20 and 30 m. Karentz plans additional studies of how ultraviolet may affect seasonal plankton blooms} (growth spurts) in the oceans.[54]
See also
References
- ↑ Turner, Anthony; Wilson, George; Kaube, Isao (1987). Biosensors:Fundamentals and Applications. Oxford, UK: Oxford University Press. p. 770. ISBN 0198547242.
- ↑ Bănică, Florinel-Gabriel (2012). Chemical Sensors and Biosensors:Fundamentals and Applications. Chichester, UK: John Wiley & Sons. p. 576. ISBN 9781118354230.
- ↑ Cavalcanti A, Shirinzadeh B, Zhang M, Kretly LC (2008). "Nanorobot Hardware Architecture for Medical Defense" (PDF). Sensors 8 (5): 2932–2958. doi:10.3390/s8052932.
- ↑ "UCSB Electronic nose, http://www.microfluidicsolutions.com/apps/blog/show/20808263-ucsb-sensor-sniffs-explosives-through-microfluidics-might-replace-rover-at-the-airport-video-". External link in
|title=(help); - ↑ "Wasp Hound". Science Central. Archived from the original on July 16, 2011. Retrieved 23 February 2011.
- 1 2 "MolluSCAN eye". MolluSCAN eye. CNRS & Université de Bordeaux. Retrieved 24 June 2015.
- ↑ S.Zeng; Baillargeat, Dominique; Ho, Ho-Pui; Yong, Ken-Tye; et al. (2014). "Nanomaterials enhanced surface plasmon resonance for biological and chemical sensing applications" (PDF). Chemical Society Reviews 43 (10): 3426–3452. doi:10.1039/C3CS60479A. PMID 24549396.
- ↑ Krupin, O., Wang, C., Berini, P. "Optical plasmonic biosensor for leukemia detection". SPIE Newsroom (22 January 2016). doi:10.1117/2.1201512.006268.
- ↑ Homola J (2003 /8x2n9xhbkqtp76dq). "Present and future of surface plasmon resonance biosensors.". Check date values in:
|date=(help) - ↑ Iqbal, M.; Gleeson, M. A.; Spaugh, B.; Tybor, F.; Gunn, W. G.; Hochberg, M.; Baehr-Jones, T.; Bailey, R. C.; Gunn, L. C. (2010). "Label-Free Biosensor Arrays Based on Silicon Ring Resonators and High-Speed Optical Scanning Instrumentation". IEEE J. Sel. Top. Quant. Elec. 16: 654–661. doi:10.1109/jstqe.2009.2032510.
- ↑ J. Witzens, M. Hochberg (2011). "Optical detection of target molecule induced aggregation of nanoparticles by means of high-Q resonators". Opt. Express 19: 7034–7061. Bibcode:2011OExpr..19.7034W. doi:10.1364/oe.19.007034.
- ↑ Hiep, H. M.; et al. (2007). "A localized surface plasmon resonance based immunosensor for the detection of casein in milk". Sci. Technol. Adv. Mater 8: 331–338. Bibcode:2007STAdM...8..331M. doi:10.1016/j.stam.2006.12.010.
- ↑ Urban, Gerald A (2009). "Micro- and nanobiosensors—state of the art and trends". Meas. Sci. Technol 20: 012001. Bibcode:2009MeScT..20a2001U. doi:10.1088/0957-0233/20/1/012001.
- ↑ Fan, F.; et al. (2008). "Novel Genetically Encoded Biosensors Using Firefly Luciferase". ACS Chem. Biol. 3: 346–51. doi:10.1021/cb8000414.
- ↑ A. Hierlemann, O. Brand, C. Hagleitner, H. Baltes, "Microfabrication techniques for chemical/biosensors, Proceedings of the IEEE 91 (6), 2003, 839-863.
- ↑ A. Hierlemann, H. Baltes, "CMOS-based chemical microsensors, The Analyst 128 (1), 2003, pp. 15-28.
- ↑ "Biosensors Primer". Retrieved 28 January 2013.
- ↑ Vo-Dinh, T.; Cullum, B. (2000). "Biosensors and biochips: Advances in biological and medical diagnostics". Fresenius' Journal of Analytical Chemistry 366 (6–7): 540–551. doi:10.1007/s002160051549.
- 1 2 3 Marazuela, M.; Moreno-Bondi, M. (2002). "Fiber-optic biosensors - an overview". Analytical and Bioanalytical Chemistry 372 (5–6): 664–682. doi:10.1007/s00216-002-1235-9. PMID 11941437.
- ↑ Pohanka, M (2013). "Cholinesterases in biorecognition and biosensor construction, a review.". Analytical Letters 46 (12): 1849–1868. doi:10.1080/00032719.2013.780240.
- ↑ Donzella, V; Crea, F (Jun 2011). "Optical biosensors to analyze novel biomarkers in oncology". J Biophotonics 4 (6): 442–52. doi:10.1002/jbio.201000123.
- ↑ Vollmer, F; Yang, Lang (Oct 2012). "Label-free detection with high-Q microcavities: a review of biosensing mechanisms for integrated devices". Nanophotonics 1: 267–291. Bibcode:2012Nanop...1..267V. doi:10.1515/nanoph-2012-0021.
- ↑ Rizzuto, R.; Pinton, P.; Brini, M.; Chiesa, A.; Filippin, L.; Pozzan, T. (1999). "Mitochondria as biosensors of calcium microdomains". Cell Calcium 26 (5): 193–199. doi:10.1054/ceca.1999.0076. PMID 10643557.
- ↑ Bragadin, M.; Manente, S.; Piazza, R.; Scutari, G. (2001). "The Mitochondria as Biosensors for the Monitoring of Detergent Compounds in Solution". Analytical Biochemistry 292 (2): 305–307. doi:10.1006/abio.2001.5097. PMID 11355867.
- ↑ Védrine, C.; Leclerc, J.-C.; Durrieu, C.; Tran-Minh, C. (2003). "Optical whole-cell biosensor using Chlorella vulgaris designed for monitoring herbicides". Biosensors & bioelectronics 18 (4): 457–63. doi:10.1016/s0956-5663(02)00157-4.
- ↑ Dubey, R. S.; Upadhyay, S. N. (2001). "Microbial corrosion monitoring by an amperometric microbial biosensor developed using whole cell of Pseudomonas sp.". Biosensors & bioelectronics 16 (9-12): 995–1000. doi:10.1016/s0956-5663(01)00203-2.
- ↑ Campàs, M.; Carpentier, R.; Rouillon, R. (2008). "Plant tissue-and photosynthesis-based biosensors". Biotechnology Advances 26 (4): 370–378. doi:10.1016/j.biotechadv.2008.04.001. PMID 18495408.
- ↑ Pickup, JC; Zhi, ZL; Khan, F; Saxl, T; Birch, DJ (2008). "Nanomedicine and its potential in diabetes research and practice". Diabetes Metab Res Rev. 24 (8): 604–10. doi:10.1002/dmrr.893. PMID 18802934.
- ↑ Gupta, R; Chaudhury, NK (May 2007). "Entrapment of biomolecules in sol-gel matrix for applications in biosensors: problems and future prospects". Biosens Bioelectron 22 (11): 2387–99. doi:10.1016/j.bios.2006.12.025. PMID 17291744.
- ↑ Clark, HA; Kopelman, R; Tjalkens, R; Philbert, MA (Nov 1999). "Optical nanosensors for chemical analysis inside single living cells. 2. Sensors for pH and calcium and the intracellular application of PEBBLE sensors". Anal Chem. 71 (21): 4837–43. doi:10.1021/ac990630n.
- ↑ Liao, KC; Hogen-Esch, T; Richmond, FJ; Marcu, L; Clifton, W; Loeb, GE (May 2008). "Percutaneous fiber-optic sensor for chronic glucose monitoring in vivo". Biosens Bioelectron 23 (10): 1458–65. doi:10.1016/j.bios.2008.01.012. PMID 18304798.
- ↑ Mimicking Body Biosensors | MIT Technology Review
- ↑ Lud, S.Q.; Nikolaides, M.G.; Haase, I.; Fischer, M.; Bausch, A.R. (2006). "Field Effect of Screened Charges: Electrical Detection of Peptides and Proteins by a Thin Film Resistor". ChemPhysChem 7 (2): 379–384. doi:10.1002/cphc.200500484.
- ↑ Vockenroth I, Atanasova P, Knoll W, Jenkins A, Köper I (2005). "Functional tethered bilayer membranes as a biosensor platform". IEEE Sensors 2005 - the 4-th IEEE Conference on Sensors: 608–610. doi:10.1109/icsens.2005.1597772.
- ↑ Cornell BA; BraachMaksvytis VLB; King LG; et al. (1997). "A biosensor that uses ion-channel switches". Nature 387 (6633): 580–583. Bibcode:1997Natur.387..580C. doi:10.1038/42432. PMID 9177344.
- ↑ Oh S; Cornell B; Smith D; et al. (2008). "Rapid detection of influenza A virus in clinical samples using an ion channel switch biosensor". Biosensors & Bioelectronics 23 (7): 1161–1165. doi:10.1016/j.bios.2007.10.011.
- ↑ Krishnamurthy V, Monfared S, Cornell B (2010). ": Ion Channel Biosensors Part I Construction Operation and Clinical Studies". IEEE Transactions on Nanotechnology 9 (3): 313–322. Bibcode:2010ITNan...9..313K. doi:10.1109/TNANO.2010.2041466.
- ↑ Kotanen, Christian N.; Gabriel Moussy, Francis; Carrara, Sandro; Guiseppi-Elie, Anthony (2012). "Implantable enzyme amperometric biosensors". Biosensors and Bioelectronics 35 (1): 14–26. doi:10.1016/j.bios.2012.03.016. PMID 22516142.
- ↑ Pasco, Neil; Glithero, Nick. Lactose at-line biosensor 1st viable industrial biosensor? http://nzbio2012.co.nz/content/nzbio2012/images/3_Biosensor_Development_for_Detecting_Lactose_in_Dairy_Wastewater__Neil_Pasco.pdf (accessed Jan 30, 2013).
- ↑ Saharudin Haron and Asim K. Ray (2006) Optical biodetection of cadmium and lead ions in water. Medical Engineering and Physics, 28 (10). pp. 978-981.
- ↑ Pohanka, M; Skladal, P; Kroca, M (2007). "Biosensors for biological warfare agent detection". Def. Sci. J. 57 (3): 185–93. doi:10.14429/dsj.57.1760.
- ↑ "Protein Engineering and Electrochemical Biosensors". Advances in Biochemical Engineering/Biotechnology: 65–96. doi:10.1007/10_2007_080.
- ↑ Pohanka, M; Jun, D; Kuca, K (2007). "Mycotoxin assay using biosensor technology: a review". Drug Chem. Toxicol 30 (3): 253–61. doi:10.1080/01480540701375232.
- ↑ Ghoshdastider U, Wu R, Trzaskowski B, Mlynarczyk K, Miszta P, Gurusaran M, Viswanathan S, Renugopalakrishnan V, Filipek S (2015). "Nano-Encapsulation of Glucose Oxidase Dimer by Graphene". RSC Advances 5 (18): 13570–78. doi:10.1039/C4RA16852F.
- ↑ Daaboul, G.G.; et al. (2010). "LED-based Interferometric Reflectance Imaging Sensor for quantitative dynamic monitoring of biomolecular interactions". Biosens. Bioelectron 26: 2221–2227. doi:10.1016/j.bios.2010.09.038.
- ↑ Ahn, S.; Freedman, D. S.; Massari, P.; Cabodi, M.; Ünlü, M. S. (2013). "A Mass-Tagging Approach for Enhanced Sensitivity of Dynamic Cytokine Detection Using a Label-Free Biosensor". Langmuir 29 (17): 5369–5376. doi:10.1021/la400982h.
- ↑ Reddington, A.; Trueb, J. T.; Freedman, D. S.; Tuysuzoglu, A.; Daaboul, G. G.; Lopez, C. A.; Karl, W. C.; Connor, J. H.; Fawcett, H. E.; Ünlü, M. S. (2013). "An Interferometric Reflectance Imaging Sensor for Point of Care Viral Diagnostics". IEEE Transactions on Biomedical Engineering 60 (12): 3276–3283. doi:10.1109/tbme.2013.2272666.
- 1 2 Monroe, M. R.; Reddington, A.; Collins, A. D.; Laboda, C. D.; Cretich, M.; Chiari, M.; Little, F. F.; Ünlü, M. S. (2011). "Multiplexed method to calibrate and quantitate fluorescence signal for allergen-specific IgE". Analytical Chemistry 83: 9485–9491. doi:10.1021/ac202212k.
- ↑ Yurt, A.; Daaboul, G. G.; Connor, J. H.; Goldberg, B. B.; Ünlü, M. S. (2012). "Single nanoparticle detectors for biological applications". Nanoscale 4 (3): 715–726. Bibcode:2012Nanos...4..715Y. doi:10.1039/c2nr11562j.
- ↑ C. A. Lopez, G. G. Daaboul, R. S. Vedula, E. Ozkumur, D. A. Bergstein, T. W. Geisbert, H. Fawcett, B. B. Goldberg, J. H. Connor, and M. S. Ünlü, "Label-free multiplexed virus detection using spectral reflectance imaging," Biosensors and Bioelectronics, 2011
- ↑ Monroe, M. R.; Daaboul, G. G.; Tuysuzoglu, A.; Lopez, C. A.; Little, F. F.; Ünlü, M. S. (2013). "Single Nanoparticle Detection for Multiplexed Protein Diagnostics with Attomolar Sensitivity in Serum and Unprocessed Whole Blood". Analytical Chemistry 85 (7): 3698–3706. doi:10.1021/ac4000514.
- ↑ Daaboul, G. G.; Yurt, A.; Zhang, X.; Hwang, G. M.; Goldberg, B. B.; Ünlü, M. S. (2010). "High-Throughput Detection and Sizing of Individual Low-Index Nanoparticles and Viruses for Pathogen Identification". Nano Letters 11: 4727–4731. doi:10.1021/nl103210p.
- ↑ Construction and use of broad host range mercury and arsenite sensor plasmids in the soil bacterium Pseudomonas fluorescens OS8 - Springer
- ↑ J. G. Black,"Principles and explorations", edition 5th.
"The Chemistry of Health." The Chemistry of Health (2006): 42-43. National Institutes of Health and National Institute of General Medical Sciences. Web. <http://www.nigms.nih.gov>
External links
- What are biosensors
- Biosensor Applications - Drug and explosives detection products from Sweden
- Scratching at the surface of biosensors - an *Instant Insight discussing how surface chemistry lets porous silicon biosensors fulfil their promise from the Royal Society of Chemistry
- Biosensor Forum - Social network for researchers and organizations
- Department of Bioelectronics, Ecole des Mines de St. Etienne
|