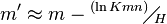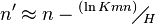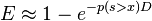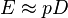BLAST
| Developer(s) | Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ, NCBI |
|---|---|
| Stable release | 2.2.31+ / 18 May 2015 |
| Operating system | UNIX, GNU/Linux, Mac, MS-Windows |
| Type | Bioinformatics tool |
| License | Public domain |
| Website |
blast |
In bioinformatics, BLAST for Basic Local Alignment Search Tool is an algorithm for comparing primary biological sequence information, such as the amino-acid sequences of different proteins or the nucleotides of DNA sequences. A BLAST search enables a researcher to compare a query sequence with a library or database of sequences, and identify library sequences that resemble the query sequence above a certain threshold.
Different types of BLASTs are available according to the query sequences. For example, following the discovery of a previously unknown gene in the mouse, a scientist will typically perform a BLAST search of the human genome to see if humans carry a similar gene; BLAST will identify sequences in the human genome that resemble the mouse gene based on similarity of sequence. The BLAST algorithm and program were designed by Stephen Altschul, Warren Gish, Webb Miller, Eugene Myers, and David J. Lipman at the National Institutes of Health and was published in the Journal of Molecular Biology in 1990 and cited over 50,000 times.[1]
Background
BLAST is one of the most widely used bioinformatics programs for sequence searching.[2] It addresses a fundamental problem in bioinformatics research. The heuristic algorithm it uses is much faster than other approaches, such as calculating an optimal alignment. This emphasis on speed is vital to making the algorithm practical on the huge genome databases currently available, although subsequent algorithms can be even faster.
Before BLAST, FASTA was developed by David J. Lipman and William R. Pearson in 1985.[3]
Before fast algorithms such as BLAST and FASTA were developed, doing database searches for protein or nucleic sequences was very time consuming because a full alignment procedure (e.g., the Smith–Waterman algorithm) was used.
While BLAST is faster than any Smith-Waterman implementation for most cases, it cannot "guarantee the optimal alignments of the query and database sequences" as Smith-Waterman algorithm does. The optimality of Smith-Waterman "ensured the best performance on accuracy and the most precise results" at the expense of time and computer power.
BLAST is more time-efficient than FASTA by searching only for the more significant patterns in the sequences, yet with comparative sensitivity. This could be further realized by understanding the algorithm of BLAST introduced below.
Examples of other questions that researchers use BLAST to answer are:
- Which bacterial species have a protein that is related in lineage to a certain protein with known amino-acid sequence?
- What other genes encode proteins that exhibit structures or motifs such as ones that have just been determined?
BLAST is also often used as part of other algorithms that require approximate sequence matching.
The BLAST algorithm and the computer program that implements it were developed by Stephen Altschul, Warren Gish, and David Lipman at the U.S. National Center for Biotechnology Information (NCBI), Webb Miller at the Pennsylvania State University, and Gene Myers at the University of Arizona. It is available on the web on the NCBI website. Alternative implementations include AB-BLAST (formerly known as WU-BLAST), FSA-BLAST (last updated in 2006), and ScalaBLAST.[4][5]
The original paper by Altschul, et al.[1] was the most highly cited paper published in the 1990s.[6]
Input
Input sequences (in FASTA or Genbank format) and weight matrix.
Output
BLAST output can be delivered in a variety of formats. These formats include HTML, plain text, and XML formatting. For NCBI's web-page, the default format for output is HTML. When performing a BLAST on NCBI, the results are given in a graphical format showing the hits found, a table showing sequence identifiers for the hits with scoring related data, as well as alignments for the sequence of interest and the hits received with corresponding BLAST scores for these. The easiest to read and most informative of these is probably the table.
If one is attempting to search for a proprietary sequence or simply one that is unavailable in databases available to the general public through sources such as NCBI, there is a BLAST program available for download to any computer, at no cost. This can be found at BLAST+ executables. There are also commercial programs available for purchase. Databases can be found from the NCBI site, as well as from Index of BLAST databases (FTP).
Process
Using a heuristic method, BLAST finds similar sequences, by locating short matches between the two sequences. This process of finding similar sequences is called seeding. It is after this first match that BLAST begins to make local alignments. While attempting to find similarity in sequences, sets of common letters, known as words, are very important. For example, suppose that the sequence contains the following stretch of letters, GLKFA. If a BLAST was being conducted under normal conditions, the word size would be 3 letters. In this case, using the given stretch of letters, the searched words would be GLK, LKF, KFA. The heuristic algorithm of BLAST locates all common three-letter words between the sequence of interest and the hit sequence or sequences from the database. This result will then be used to build an alignment. After making words for the sequence of interest, the rest of the words are also assembled. These words must satisfy a requirement of having a score of at least the threshold T, when compared by using a scoring matrix. One commonly-used scoring matrix for BLAST searches is BLOSUM62, although the optimal scoring matrix depends on sequence similarity. Once both words and neighborhood words are assembled and compiled, they are compared to the sequences in the database in order to find matches. The threshold score T determines whether or not a particular word will be included in the alignment. Once seeding has been conducted, the alignment which is only 3 residues long, is extended in both directions by the algorithm used by BLAST. Each extension impacts the score of the alignment by either increasing or decreasing it. If this score is higher than a pre-determined T, the alignment will be included in the results given by BLAST. However, if this score is lower than this pre-determined T, the alignment will cease to extend, preventing the areas of poor alignment from being included in the BLAST results. Note that increasing the T score limits the amount of space available to search, decreasing the number of neighborhood words, while at the same time speeding up the process of BLAST.
Algorithm
To run the software, BLAST requires a query sequence to search for, and a sequence to search against (also called the target sequence) or a sequence database containing multiple such sequences. BLAST will find sub-sequences in the database which are similar to sub sequences in the query. In typical usage, the query sequence is much smaller than the database, e.g., the query may be one thousand nucleotides while the database is several billion nucleotides.
The main idea of BLAST is that there are often High-scoring Segment Pairs (HSP) contained in a statistically significant alignment. BLAST searches for high scoring sequence alignments between the query sequence and the existing sequences in the database using a heuristic approach that approximates the Smith-Waterman algorithm. However, the exhaustive Smith-Waterman approach is too slow for searching large genomic databases such as GenBank. Therefore, the BLAST algorithm uses a heuristic approach that is less accurate than the Smith-Waterman algorithm but over 50 times faster. [8] The speed and relatively good accuracy of BLAST are among the key technical innovations of the BLAST programs.
An overview of the BLAST algorithm (a protein to protein search) is as follows:[7]
- Remove low-complexity region or sequence repeats in the query sequence.
- "Low-complexity region" means a region of a sequence composed of few kinds of elements. These regions might give high scores that confuse the program to find the actual significant sequences in the database, so they should be filtered out. The regions will be marked with an X (protein sequences) or N (nucleic acid sequences) and then be ignored by the BLAST program. To filter out the low-complexity regions, the SEG program is used for protein sequences and the program DUST is used for DNA sequences. On the other hand, the program XNU is used to mask off the tandem repeats in protein sequences.
- Make a k-letter word list of the query sequence.
- Take k=3 for example, we list the words of length 3 in the query protein sequence (k is usually 11 for a DNA sequence) "sequentially", until the last letter of the query sequence is included. The method is illustrated in figure 1.

- Take k=3 for example, we list the words of length 3 in the query protein sequence (k is usually 11 for a DNA sequence) "sequentially", until the last letter of the query sequence is included. The method is illustrated in figure 1.
- List the possible matching words.
- This step is one of the main differences between BLAST and FASTA. FASTA cares about all of the common words in the database and query sequences that are listed in step 2; however, BLAST only cares about the high-scoring words. The scores are created by comparing the word in the list in step 2 with all the 3-letter words. By using the scoring matrix (substitution matrix) to score the comparison of each residue pair, there are 20^3 possible match scores for a 3-letter word. For example, the score obtained by comparing PQG with PEG and PQA is 15 and 12, respectively. For DNA words, a match is scored as +5 and a mismatch as -4, or as +2 and -3. After that, a neighborhood word score threshold T is used to reduce the number of possible matching words. The words whose scores are greater than the threshold T will remain in the possible matching words list, while those with lower scores will be discarded. For example, PEG is kept, but PQA is abandoned when T is 13.
- Organize the remaining high-scoring words into an efficient search tree.
- This allows the program to rapidly compare the high-scoring words to the database sequences.
- Repeat step 3 to 4 for each k-letter word in the query sequence.
- Scan the database sequences for exact matches with the remaining high-scoring words.
- The BLAST program scans the database sequences for the remaining high-scoring word, such as PEG, of each position. If an exact match is found, this match is used to seed a possible un-gapped alignment between the query and database sequences.
- Extend the exact matches to high-scoring segment pair (HSP).
- The original version of BLAST stretches a longer alignment between the query and the database sequence in the left and right directions, from the position where the exact match occurred. The extension does not stop until the accumulated total score of the HSP begins to decrease. A simplified example is presented in figure 2.


- To save more time, a newer version of BLAST, called BLAST2 or gapped BLAST, has been developed. BLAST2 adopts a lower neighborhood word score threshold to maintain the same level of sensitivity for detecting sequence similarity. Therefore, the possible matching words list in step 3 becomes longer. Next, the exact matched regions, within distance A from each other on the same diagonal in figure 3, will be joined as a longer new region. Finally, the new regions are then extended by the same method as in the original version of BLAST, and the HSPs' (High-scoring segment pair) scores of the extended regions are then created by using a substitution matrix as before.
- The original version of BLAST stretches a longer alignment between the query and the database sequence in the left and right directions, from the position where the exact match occurred. The extension does not stop until the accumulated total score of the HSP begins to decrease. A simplified example is presented in figure 2.
- List all of the HSPs in the database whose score is high enough to be considered.
- We list the HSPs whose scores are greater than the empirically determined cutoff score S. By examining the distribution of the alignment scores modeled by comparing random sequences, a cutoff score S can be determined such that its value is large enough to guarantee the significance of the remaining HSPs.
- Evaluate the significance of the HSP score.
- BLAST next assesses the statistical significance of each HSP score by exploiting the Gumbel extreme value distribution (EVD). (It is proved that the distribution of Smith-Waterman local alignment scores between two random sequences follows the Gumbel EVD. For local alignments containing gaps it is not proved.). In accordance with the Gumbel EVD, the probability p of observing a score S equal to or greater than x is given by the equation
-
- where
-
- The statistical parameters
 and
and 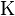 are estimated by fitting the distribution of the un-gapped local alignment scores, of the query sequence and a lot of shuffled versions (Global or local shuffling) of a database sequence, to the Gumbel extreme value distribution. Note that
are estimated by fitting the distribution of the un-gapped local alignment scores, of the query sequence and a lot of shuffled versions (Global or local shuffling) of a database sequence, to the Gumbel extreme value distribution. Note that  and
and 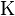 depend upon the substitution matrix, gap penalties, and sequence composition (the letter frequencies).
depend upon the substitution matrix, gap penalties, and sequence composition (the letter frequencies).  and
and  are the effective lengths of the query and database sequences, respectively. The original sequence length is shortened to the effective length to compensate for the edge effect (an alignment start near the end of one of the query or database sequence is likely not to have enough sequence to build an optimal alignment). They can be calculated as
are the effective lengths of the query and database sequences, respectively. The original sequence length is shortened to the effective length to compensate for the edge effect (an alignment start near the end of one of the query or database sequence is likely not to have enough sequence to build an optimal alignment). They can be calculated as
-
- where
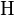 is the average expected score per aligned pair of residues in an alignment of two random sequences. Altschul and Gish gave the typical values,
is the average expected score per aligned pair of residues in an alignment of two random sequences. Altschul and Gish gave the typical values, 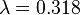 ,
, 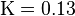 , and
, and 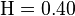 , for un-gapped local alignment using BLOSUM62 as the substitution matrix. Using the typical values for assessing the significance is called the lookup table method; it is not accurate. The expect score E of a database match is the number of times that an unrelated database sequence would obtain a score S higher than x by chance. The expectation E obtained in a search for a database of D sequences is given by
, for un-gapped local alignment using BLOSUM62 as the substitution matrix. Using the typical values for assessing the significance is called the lookup table method; it is not accurate. The expect score E of a database match is the number of times that an unrelated database sequence would obtain a score S higher than x by chance. The expectation E obtained in a search for a database of D sequences is given by
-
- Furthermore, when
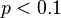 , E could be approximated by the Poisson distribution as
, E could be approximated by the Poisson distribution as
-
- This expectation or expect value "E" (often called an E score or E-value or e-value) assessing the significance of the HSP score for un-gapped local alignment is reported in the BLAST results. The calculation shown here is modified if individual HSPs are combined, such as when producing gapped alignments (described below), due to the variation of the statistical parameters.
- BLAST next assesses the statistical significance of each HSP score by exploiting the Gumbel extreme value distribution (EVD). (It is proved that the distribution of Smith-Waterman local alignment scores between two random sequences follows the Gumbel EVD. For local alignments containing gaps it is not proved.). In accordance with the Gumbel EVD, the probability p of observing a score S equal to or greater than x is given by the equation
- Make two or more HSP regions into a longer alignment.
- Sometimes, we find two or more HSP regions in one database sequence that can be made into a longer alignment. This provides additional evidence of the relation between the query and database sequence. There are two methods, the Poisson method and the sum-of-scores method, to compare the significance of the newly combined HSP regions. Suppose that there are two combined HSP regions with the pairs of scores (65, 40) and (52, 45), respectively. The Poisson method gives more significance to the set with the maximal lower score (45>40). However, the sum-of-scores method prefers the first set, because 65+40 (105) is greater than 52+45(97). The original BLAST uses the Poisson method; gapped BLAST and the WU-BLAST uses the sum-of scores method.
- Show the gapped Smith-Waterman local alignments of the query and each of the matched database sequences.
- The original BLAST only generates un-gapped alignments including the initially found HSPs individually, even when there is more than one HSP found in one database sequence.
- BLAST2 produces a single alignment with gaps that can include all of the initially-found HSP regions. Note that the computation of the score and its corresponding E-value involves use of adequate gap penalties.
- Report every match whose expect score is lower than a threshold parameter E.
Parallel BLAST
Parallel BLAST versions of split databases are implemented using MPI and Pthreads, and have been ported to various platforms including Windows, Linux, Solaris, Mac OS X, and AIX. Popular approaches to parallelize BLAST include query distribution, hash table segmentation, computation parallelization, and database segmentation (parttion). Databases are split into equal sized pieces and stored locally on each node. Each query is run on all nodes in parallel and the resultant BLAST output files from all nodes merged to yield the final output.[3]
Program
The BLAST program can either be downloaded and run as a command-line utility "blastall" or accessed for free over the web. The BLAST web server, hosted by the NCBI, allows anyone with a web browser to perform similarity searches against constantly updated databases of proteins and DNA that include most of the newly sequenced organisms.
The BLAST program is based on an open-source format, giving everyone access to it and enabling them to have the ability to change the program code. This has led to the creation of several BLAST "spin-offs".
There are now a handful of different BLAST programs available, which can be used depending on what one is attempting to do and what they are working with. These different programs vary in query sequence input, the database being searched, and what is being compared. These programs and their details are listed below:
BLAST is actually a family of programs (all included in the blastall executable). These include:[8]
- Nucleotide-nucleotide BLAST (blastn)
- This program, given a DNA query, returns the most similar DNA sequences from the DNA database that the user specifies.
- Protein-protein BLAST (blastp)
- This program, given a protein query, returns the most similar protein sequences from the protein database that the user specifies.
- Position-Specific Iterative BLAST (PSI-BLAST) (blastpgp)
- This program is used to find distant relatives of a protein. First, a list of all closely related proteins is created. These proteins are combined into a general "profile" sequence, which summarises significant features present in these sequences. A query against the protein database is then run using this profile, and a larger group of proteins is found. This larger group is used to construct another profile, and the process is repeated.
- By including related proteins in the search, PSI-BLAST is much more sensitive in picking up distant evolutionary relationships than a standard protein-protein BLAST.
- Nucleotide 6-frame translation-protein (blastx)
- This program compares the six-frame conceptual translation products of a nucleotide query sequence (both strands) against a protein sequence database.
- Nucleotide 6-frame translation-nucleotide 6-frame translation (tblastx)
- This program is the slowest of the BLAST family. It translates the query nucleotide sequence in all six possible frames and compares it against the six-frame translations of a nucleotide sequence database. The purpose of tblastx is to find very distant relationships between nucleotide sequences.
- Protein-nucleotide 6-frame translation (tblastn)
- This program compares a protein query against the all six reading frames of a nucleotide sequence database.
- Large numbers of query sequences (megablast)
- When comparing large numbers of input sequences via the command-line BLAST, "megablast" is much faster than running BLAST multiple times. It concatenates many input sequences together to form a large sequence before searching the BLAST database, then post-analyzes the search results to glean individual alignments and statistical values.
Of these programs, BLASTn and BLASTp are the most commonly used because they use direct comparisons, and do not require translations. However, since protein sequences are better conserved evolutionarily than nucleotide sequences, tBLASTn, tBLASTx, and BLASTx, produce more reliable and accurate results when dealing with coding DNA. They also enable one to be able to directly see the function of the protein sequence, since by translating the sequence of interest before searching often gives you annotated protein hits.
Alternative versions
A version designed for comparing large genomes or DNA is BLASTZ.
CS-BLAST (ContSxt-Specific BLAST) is an extended version of BLAST for searching protein sequences that finds twice as many remotely related sequences as BLAST at the same speed and error rate. In CS-BLAST, the mutation probabilities between amino acids depend not only on the single amino acid, as in BLAST, but also on its local sequence context.Washington University produced an alternative version of NCBI BLAST, called WU-BLAST. The rights have since been acquired to Advanced Biocomputing, LLC.
In 2009, NCBI has released a new set of BLAST executables, the C++ based BLAST+,[9] and has released parallel versions until 2.2.26. Starting with version 2.2.27 (April 2013), only BLAST+ executables are available. Among the changes is the replacement of the blastall executable with separate executables for the different BLAST programs, and changes in option handling. The formatdb utility (C based) has been replaced by makeblastdb (C++ based) and databases formatted by either one should be compatible for identical blast releases. The algorithms remain similar, however, the number of hits found and their order can vary significantly between the older and the newer version.
Accelerated versions
- CLC bio and SciEngines GmbH collaborate on an FPGA accelerator they claim will give 188x acceleration of BLAST.
- TimeLogic offers another FPGA-accelerated implementation of the BLAST algorithm called Tera-BLAST.
- The Mitrion-C Open Bio Project is an ongoing effort to port BLAST to run on Mitrion FPGAs.
- The GPU-Blast is an accelerated version of NCBI BLASTP for CUDA which is 3x~4x faster than NCBI Blast.[10]
- The CUDA-BLASTP is a version of BLASTP that is GPU-accelerated and is claimed to run up to 10x faster than NCBI BLAST.
- G-BLASTN is an accelerated version of NCBI blastn and megablast, whose speedup varies from 4x to 14x (compared to the same runs with 4 CPU threads).[11] Its current limitation is that the database must fit into the GPU memory.
- MPIBlast is a parallel implementation of NCBI BLAST using Message Passing Interface. By efficiently utilizing distributed computational resources through database fragmentation, query segmentation, intelligent scheduling, and parallel I/O, mpiBLAST improves NCBI BLAST performance by several orders of magnitude while scaling to hundreds of processors.
- Paracel BLAST is a commercial parallel implementation of NCBI BLAST, supporting hundreds of processors.
- CaBLAST makes search on large databases orders of magnitude faster by exploiting redundancy in data.
Alternatives to BLAST
An extremely fast but considerably less sensitive alternative to BLAST is BLAT (Blast Like Alignment Tool). While BLAST does a linear search, BLAT relies on k-mer indexing the database, and can thus often find seeds faster. Another software alternative similar to BLAT is PatternHunter.
Advances in sequencing technology in the late 2000s has made searching for very similar nucleotide matches an important problem. New alignment programs tailored for this use typically use BWT-indexing of the target database (typically a genome). Input sequences can then be mapped very quickly, and output is typically in the form of a BAM file. Example alignment programs are BWA, SOAP, and Bowtie.
For protein identification, searching for known domains (for instance from Pfam) by matching with Hidden Markov Models is a popular alternative, such as HMMER.
An alternative to BLAST for comparing two banks of sequences is KLAST. KLAST provides a high-performance general purpose bank to bank sequence similarity search tool relying on PLAST[12] and ORIS[13] algorithms. Results of KLAST are very similar to BLAST, but KLAST is significantly faster and capable of comparing large sets of sequences with a small memory (i.e. RAM) footprint.
For applications in metagenomics, where the task is to compare billions of short DNA reads against tens of millions of protein references, DIAMOND[14] runs at up to 20,000 times as fast as BLASTX, while maintaining a high level of sensitivity.
Uses of BLAST
BLAST can be used for several purposes. These include identifying species, locating domains, establishing phylogeny, DNA mapping, and comparison.
- Identifying species
- With the use of BLAST, you can possibly correctly identify a species or find homologous species. This can be useful, for example, when you are working with a DNA sequence from an unknown species.
- Locating domains
- When working with a protein sequence you can input it into BLAST, to locate known domains within the sequence of interest.
- Establishing phylogeny
- Using the results received through BLAST you can create a phylogenetic tree using the BLAST web-page. Phylogenies based on BLAST alone are less reliable than other purpose-built computational phylogenetic methods, so should only be relied upon for "first pass" phylogenetic analyses.
- DNA mapping
- When working with a known species, and looking to sequence a gene at an unknown location, BLAST can compare the chromosomal position of the sequence of interest, to relevant sequences in the database(s).
- Comparison
- When working with genes, BLAST can locate common genes in two related species, and can be used to map annotations from one organism to another.
Comparing BLAST and the Smith-Waterman Process
While both Smith-Waterman and BLAST are used to find homologous sequences by searching and comparing a query sequence with those in the databases, they do have their differences.
Due to the fact that BLAST is based on a heuristic algorithm, the results received through BLAST, in terms of the hits found, may not be the best possible results, as it will not provide you with all the hits within the database. BLAST misses hard to find matches.
A better alternative in order to find the best possible results would be to use the Smith-Waterman algorithm. This method varies from the BLAST method in two areas, accuracy and speed. The Smith-Waterman option provides better accuracy, in that it finds matches that BLAST cannot, because it does not miss any information. Therefore, it is necessary for remote homology. However, when compared to BLAST, it is more time consuming, not to mention that it requires large amounts of computer usage and space. However, technologies to speed up the Smith-Waterman process have been found to improve the time necessary to perform a search dramatically. These technologies include FPGA chips and SIMD technology.
In order to receive better results from BLAST, the settings can be changed from their default settings. However, there is no given or set way of changing these settings in order to receive the best results for a given sequence. The settings available for change are E-Value, gap costs, filters, word size, and substitution matrix. Note, that the algorithm used for BLAST was developed from the algorithm used for Smith-Waterman. BLAST employs an alignment which finds "local alignments between sequences by finding short matches and from these initial matches (local) alignments are created".
See also
References
- 1 2 Altschul, Stephen; Gish, Warren; Miller, Webb; Myers, Eugene; Lipman, David (1990). "Basic local alignment search tool". Journal of Molecular Biology 215 (3): 403–410. doi:10.1016/S0022-2836(05)80360-2. PMID 2231712.
- ↑ Casey, R. M. (2005). "BLAST Sequences Aid in Genomics and Proteomics". Business Intelligence Network.
- ↑ Lipman, DJ; Pearson, WR (1985). "Rapid and sensitive protein similarity searches". Science 227 (4693): 1435–41. doi:10.1126/science.2983426. PMID 2983426.
- ↑ Oehmen, C.; Nieplocha, J. (2006). "ScalaBLAST: A Scalable Implementation of BLAST for High-Performance Data-Intensive Bioinformatics Analysis". IEEE Transactions on Parallel and Distributed Systems 17 (8): 740. doi:10.1109/TPDS.2006.112.
- ↑ Oehmen, C. S.; Baxter, D. J. (2013). "ScalaBLAST 2.0: Rapid and robust BLAST calculations on multiprocessor systems". Bioinformatics 29 (6): 797–798. doi:10.1093/bioinformatics/btt013. PMC 3597145. PMID 23361326.
- ↑ "Sense from Sequences: Stephen F. Altschul on Bettering BLAST". ScienceWatch. July–August 2000. Archived from the original on October 7, 2007.
- ↑ Mount, D. W. (2004). Bioinformatics: Sequence and Genome Analysis (2nd ed.). Cold Spring Harbor Press. ISBN 978-0-87969-712-9.
- ↑ "Program Selection Tables of the Blast NCBI web site".
- ↑ Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T. L. (2009). "BLAST+: Architecture and applications". BMC Bioinformatics 10: 421. doi:10.1186/1471-2105-10-421. PMC 2803857. PMID 20003500.
- ↑ Vouzis, P. D.; Sahinidis, N. V. (2010). "GPU-BLAST: using graphics processors to accelerate protein sequence alignment". Bioinformatics 27 (2): 182–8. doi:10.1093/bioinformatics/btq644. PMC 3018811. PMID 21088027.
- ↑ Zhao, K.; Chu, X. (2014). "G-BLASTN: accelerating nucleotide alignment by graphics processors". Bioinformatics 30 (10): 1384–91. doi:10.1093/bioinformatics/btu047. PMID 24463183.
- ↑ Lavenier, D.; Lavenier, Dominique (2009). "PLAST: parallel local alignment search tool for database comparison". BMC Bioinformatics 10: 329. doi:10.1186/1471-2105-10-329. PMC 2770072. PMID 19821978.
- ↑ Lavenier, D. (2009). "Ordered index seed algorithm for intensive DNA sequence comparison". 2008 IEEE International Symposium on Parallel and Distributed Processing (PDF). p. 1. doi:10.1109/IPDPS.2008.4536172. ISBN 978-1-4244-1693-6.
- ↑ Buchfink, Xie and Huson (2015). "Fast and sensitive protein alignment using DIAMOND". Nature Methods 12: 59–60. doi:10.1038/nmeth.3176.
External links
| Library resources about Sequence alignment |
- Official website
- BLAST+ executables — free source downloads
- Biological Sequence Analysis I: Andy Baxevanis' lecture from NHGRI's Current Topics in Genome Analysis series, covering contemporary areas in genomics and bioinformatics
- What's behind BLAST?: talk by Gene Myers (slides and video)
Tutorials
- Wheeler, David; Bhagwat, Medha (2007). "Chapter 9: BLAST QuickStart". In Bergman, Nicholas H. Comparative Genomics Volumes 1 and 2. Methods in Molecular Biology. 395-396. Totowa, NJ: Humana Press. PMID 21250292.
- Mount DW (1 Jul 2007). "Using the Basic Local Alignment Search Tool (BLAST)". Cold Spring Harbor Protocols 2007 (14): pdb.top17. doi:10.1101/pdb.top17. PMID 21357135.
| ||||||||||||||||||||||
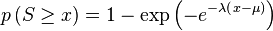
![\mu ={}^{\left[ \log \left( Km'n' \right) \right]}\!\!\diagup\!\!{}_{\lambda }\;](../I/m/de316fed5dc480dfc40e517108c01d40.png)