Asymmetric cell division
An asymmetric cell division produces two daughter cells with different cellular fates. This is in contrast to normal, symmetric, cell divisions, which give rise to daughter cells of equivalent fates. Notably, stem cells divide asymmetrically to give rise to two distinct daughter cells: one copy of the original stem cell as well as a second daughter programmed to differentiate into a non-stem cell fate. (In times of growth or regeneration, stem cells can also divide symmetrically, to produce two identical copies of the original cell.[1])
In principle, there are two mechanisms by which distinct properties may be conferred on the daughters of a dividing cell. In one, the daughter cells are initially equivalent but a difference is induced by signaling between the cells, from surrounding cells, or from the precursor cell. This mechanism is known as extrinsic asymmetric cell division. In the second mechanism, the prospective daughter cells are inherently different at the time of division of the mother cell. Because this latter mechanism does not depend on interactions of cells with each other or with their environment, it must rely on intrinsic asymmetry. The term asymmetric cell division usually refers to such intrinsic asymmetric divisions [2]
Intrinsic asymmetry
Intrinsic asymmetric divisions rely on the following mechanism. At mitosis certain proteins, RNA transcripts, and other macromolecules are localized asymmetrically to one half of the cell. A cell can accomplish this through a variety of processes such as localized molecule tethering as well as molecule transport (see below). Following this, the cell performs cytokinesis and divides in two. Thus, the asymmetrically localized proteins, RNA transcripts, and other macromolecules are inherited differentially to only one of the daughter cells, causing that cell to assume a separate fate from its sibling. Because these molecules ultimately determine the identity of the daughter cell they are called cell fate determinants.
This mechanism raises two requirements: first, the mother cell must be polarized; second, the mitotic spindle must be aligned with the axis of polarity. The cell biology of these events has been most traditionally studied in three animal models: the mouse, the nematode Caenorhabditis elegans and the fruitfly Drosophila melanogaster. Recent work in spiralian development has also discovered insightful mechanisms of asymmetric cell division
Asymmetric cell division in C. elegans
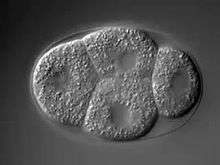
In C. elegans, a series of asymmetric cell divisions in the early embryo are critical in setting up the anterior/posterior, dorsal/ventral, and left/right axes of the body plan.[3] After fertilization, events are already occurring in the one cell stage embryo to allow for the first asymmetric cell division. This first division produces two distinctly different blastomeres, termed AB and P1. When the sperm fertilizes the egg, the sperm nucleus and centrosomes are deposited within the egg, which causes a cytoplasmic flux resulting in the movement of the sperm pronucleus and centrosomes towards one pole.[4] The centrosomes deposited by the sperm seem to be responsible for the establishment of the posterior pole within the one cell embryo. Studies have shown that the pole in which the sperm-derived centrosomes reside always becomes the posterior pole.[5] Furthermore, sperm with mutant or absent centrosomes fail to establish a posterior pole, while enucleated sperm with intact centrosomes successfully fertilize the egg and set up the posterior pole.[6][7][8] The establishment of this polarity initiates the polarized distribution of a group of proteins present in the zygote called the PAR proteins (partitioning-defective), which are a conserved group of proteins that function in establishing cell polarity during development.[9] These proteins are initially distributed uniformly throughout the zygote and then become polarized with the creation of the posterior pole. This series of events allows the single celled zygote to obtain polarity through an unequal distribution of multiple factors.
The single cell is now set up to undergo an asymmetric cell division, however the orientation in which the division occurs is also an important factor. The mitotic spindle must be oriented correctly to ensure that the proper cell fate determinants are distributed appropriately to the daughter cells. The alignment of the spindle is mediated by the PAR proteins, which regulate the positioning of the centrosomes along the A/P axis as well as the movement of the mitotic spindle along the A/P axis.[10] Following this first asymmetric division, the AB daughter cell divides symmetrically, giving rise to ABa and ABp, while the P1 daughter cell undergoes another asymmetric cell division to produce P2 and EMS. This division is also dependent on the distribution of the PAR proteins.[11]
Asymmetric cell division of Drosophila neuroblasts

In Drosophila melanogaster, asymmetric cell division plays an important role in neural development. Neuroblasts are the progenitor cells which divide asymmetrically to give rise to another neuroblast and a ganglion mother cell (GMC). The neuroblast repeatedly undergoes this asymmetric cell division while the GMC continues on to produce a pair of neurons. Two proteins play an important role in setting up this asymmetry in the neuroblast, Prospero and Numb. These proteins are both synthesized in the neuroblast and segregate into only the GMC during divisions.[12] Numb is a suppressor of Notch, therefore the asymmetric segregation of Numb to the basal cortex biases the response of the daughter cells to Notch signaling, resulting in two distinct cell fates.[13] Prospero is required for gene regulation in GMCs. It is equally distributed throughout the neuroblast cytoplasm, but becomes localized at the basal cortex when the neuroblast starts to undergo mitosis. Once the GMC buds off from the basal cortex, Prospero becomes translocated into the GMC nucleus to act as a transcription factor.[12]
Other proteins present in the neuroblast mediate the asymmetric localization of Numb and Prospero. Miranda is an anchoring protein that binds to Prospero and keeps it in the basal cortex. Following the generation of the GMC, Miranda releases Prospero and then becomes degraded.[12][14] The segregation of Numb is mediated by Pon (the partner of Numb protein). Pon binds to Numb and colocalizes with it during neuroblast cell division.[12]
The mitotic spindle must also align parallel to the asymmetrically distributed cell fate determinants to allow them to become segregated into one daughter cell and not the other. The mitotic spindle orientation is mediated by Inscuteable, which is segregated to the apical cortex of the neuroblast. Without the presence of Inscuteable, the positioning of the mitotic spindle and the cell fate determinants in relationship to each other becomes randomized. Inscuteable mutants display a uniform distribution of Miranda and Numb at the cortex, and the resulting daughter cells display identical neuronal fates.[12]
Asymmetric cell division in Spiralian development
Spiralia (commonly synonymous with lophotrochozoa) represent a diverse clade of animals whose species comprise the bulk of the bilaterian animals present today. Examples include mollusks, annelid worms, and the entoprocta. Although much is known at the cellular and molecular level about the other bilateralian clades (ecdysozoa and deuterostomia), research into the processes that govern spiralian development is comparatively lacking. However, one unifying feature shared among spiralia is the pattern of cleavage in the early embryo known as spiral cleavage.[15] This pattern, which contributed to the initial phylogenetic placement of this group of organisms, depends heavily on asymmetric cell division.
The general cleavage pattern of a spiralian embryo follows a classic and predictable set of cellular divisions. The first two divisions yield four progenitor macromeres which specify four geometric quadrants of the developing organism. Following this, the individual macromeres undergo a series of divisions which generate a string of micromeres at the animal pole of the embryo. Micromere production occurs in an alternating clockwise and counterclockwise fashion and is accomplished via asymmetric division.[15] For more information on this pattern of cellular division see the cleavage (embryo) page.
Although the conserved cleavage of spiralian macromeres to generate micromere quartets is well established, more has been discovered about the first two cleavages at the molecular level. The initial cleavage can occur with several, sometimes overlapping, outcomes (See Figure, left panel). For example, in symmetrically cleaving embryos, the zygote can cleave twice to yield four cells of equal size and equipotent fate.[15] In contrast, asymmetrically cleaving spiralians develop an embryonic polarity beginning with the first cellular division. The zygote cleaves once to generate a smaller AB cell and a larger CD cell. The second division usually occurs asynchronously, and generates three similarly sized macromeres (A, B, and C) as well as a larger D macromere. Known as D quadrant specification, this example of asymmetric cell division sets up the D quadrant of the embryo such that it has a specific and independent fate, often for mesoderm production.[15]
Mechanisms of asymmetric division (See Figure, right panel):

- Tubifex tubifex: The sludge worm Tubifex tubifex has been shown to demonstrate an interesting asymmetric cell division at the point of first embryonic cleavage. Unlike the classic idea of cortical differences at the zygotic membrane that determine spindle asymmetry in the C. elegans embryo, the first cleavage in tubifex relies on the number of centrosomes.[16] Embryos inherit a single centrosome which localizes in the prospective larger CD cell cytoplasm and emits radial microtubules during anaphase that contribute to both the mitotic spindle as well as cortical asters. However, the microtubule organizing center of the prospective smaller AB cell emits only microtubules that commit to the mitotic spindle and not cortical bound asters. When embryos are compressed or deformed, asymmetric spindles still form, and staining for gamma tubulin reveals that the second microtubule organizing center lacks the molecular signature of a centrosome. Furthermore, when centrosome number is doubled, tubifex embryos cleave symmetrically, suggesting this monoastral mechanism of asymmetric cell division is centrosome dependent.[16]
- Helobdella robusta: The leech Helobdella robusta exhibits a similar asymmetry in the first embryonic division as C. elegans and tubifex, but relies on a modified mechanism. Compression experiments on the robusta embryo do not affect asymmetric division, suggesting the mechanism, like tubifex, uses a cortical independent molecular pathway. In robusta, antibody staining reveals that the mitotic spindle forms symmetrically until metaphase and stems from two biastral centrosomes.[17] At the onset of metaphase, asymmetry becomes apparent as the centrosome of the prospective larger CD cell lengthens cortical asters while the asters of the prospective smaller AB cell become downregulated. Experiments using nocodazole and taxol support this observation. Taxol, which stabilized microtubules, forced a significant number of embryos to cleave symmetrically when used at a moderate concentration. Moreover, embryos treated with nocodazole, which sequesters tubulin dimers and promotes microtubule depolymerization, similarly forced symmetric division in a significant number of embryos. Treatment with either drug at these concentrations fails to disrupt normal centrosome dynamics, suggesting that a balance of microtubule polymerization and depolymerization represents another mechanism for establishing asymmetric cell division in spilarian development.[17]
- Ilyanasa obsoleta: A third, less traditional mechanism contributing to asymmetric cell division in spiralian development has been discovered in the mollusk Ilyanasa obsoleta. In situ hybridization and immunofluorescence experiments show that mRNA transcripts co-localize with centrosomes during early cleavage.[18] Consequently, these transcripts are inherited in a stereotypical fashion to distinct cells. All mRNA transcripts followed have been implicated in body axis patterning, and in situ hybridization for transcripts associated with other functions fail to exhibit such a localization. Moreover, disruption of microtubule polymerization with nocodazole, and of actin polymerization with cytochalisin B, shows the cytoskeleton is also important in this asymmetry. It appears that microtubules are required to recruit the mRNA to the centrosome, and that actin is required to attach the centrosome to the cortex. Finally, introducing multiple centrosomes into one cell by inhibiting cytokinesis shows that mRNA dependably localizes on the correct centrosome, suggesting intrinsic differences between each centrosomal composition. It is important to note that these results reflect experiments performed after the first two divisions, yet still demonstrate a different molecular means of establishing asymmetry in a dividing cell.[18]
These examples of mechanisms that establish asymmetric cell division in early spiralian development showcase the plasticity of evolution. A variety of methods can contribute to differences in daughter cells in order to found independent cell fates.
Asymmetric division in development
Animals are made up of a vast number of distinct cell types. During development, the zygote undergoes many cell divisions that give rise to various cell types, including embryonic stem cells. Asymmetric divisions of these embryonic cells gives rise to one cell of the same potency (self-renewal), and another that maybe of the same potency or stimulated to further differentiate into specialized cell types such as neurons. This stimulated differentiation arises from many factors which can be divided into two broad categories: intrinsic and extrinsic. Intrinsic factors generally involve differing amounts of cell-fate determinants being distributed into each daughter cell. Extrinsic factors involve interactions with neighboring cells and the micro and macro environment of the precursor cell.[19]
In addition to the aforementioned Drosophila neuronal example, it was proposed that the macrosensory organs of the Drosophila, specifically the glial cells, also arise from a similar set of asymmetric division from a single progenitor cell via regulation of the Notch signaling pathway and transcription factors.[20] The precursor cell (pI) divides to produce daughter cells (pII) which further divide to produce further cells (pIII). It was found that there is an inhibition of the Notch signaling pathway in the pIIb (the anterior daughter cell) while there is an upregulation in the pIIa (the posterior daughter cell). The pIIb is then able to asymmetrically divide to further give a subepithelial cell and a daughter cell that differentiates into glial cells.
An example of how extrinsic factors bring about this phenomenon is the physical displacement of one of the daughter cells out of the original stem cell niche, exposing it to signalling molecules such as chondroitin sulfate.[21] In this manner, the daughter cell is forced to interact with the heavily sulfated molecules, which stimulate it to differentiate while the other daughter cell remains in the original niche in a quiescent state.
Asymmetric division of stem cells plays a key role in development by allowing for the differentiation of a subset of daughter cells while maintaining stem cell pluripotency. Since it can be controlled by both intrinsic and extrinsic factors, upon delineating these particular factors it may be possible to use this knowledge in applications of tissue and whole organ generation.
Role of asymmetric cell division in disease
Asymmetric cell division is suspected to play an important role in cancer, in particular with respect to the cancer stem cell hypothesis. The hypothesis states in essence that each tumor contains a relatively small population of cells capable of initiating and maintaining tumour growth. This hypothesis has enormous therapeutic implications, but also raises the possibility that defects in stem cell lineages may lead to tumor formation. Cancer stem cells as well as normal embryonic and adult stem cells are defined by both their ability to make more stem cells, a property known as self-renewal, and their ability to produce cells that differentiate. One strategy by which cancer stem cells can accomplish these two tasks is asymmetric cell division.[1] Asymmetric division is a key mechanism to ensure tissue homeostasis.
In normal stem and progenitor cells, asymmetric cell division balances proliferation and self-renewal with cell-cycle exit and differentiation. Disruption of asymmetric cell division leads to aberrant self-renewal and impairs differentiation, and could therefore constitute an early step in the tumorogenic transformation of stem and progenitor cells. In normal non-tumor stem cells, a number of genes have been described which are responsible for pluripotency, such as Bmi-1, Wnt and Notch. These genes have been discovered also in the case of cancer stem cells, and shows that their aberrant expression is essential for the formation of tumor cell mass.[22] For example, it has been shown that gastrointestinal cancers contain rare subpopulation of cancer stem cells which are capable to divide asymmetrically. The asymmetric division in these cells is regulated by cancer niche (microenvironment) and Wnt pathway. Blocking the Wnt pathway with IWP2 (WNT antagonist) or siRNA-TCF4 resulted in high suppression of asymmetric cell division.[23]
Another mutation in asymmetric cell divisions which are involved in tumor growth are loss-of-function mutations. The first suggestion that loss of asymmetric cell division might be involved in tumorigenesis came from studies of Drosophila. Studies of loss-of-function mutations in key regulators of asymmetric cell division including lgl, aurA, polo, numb and brat, revealed hyperproliferative phenotypes in situ. In these mutants cells divide more symmetrically and generate mis-specified progeny that fail to exit the cell cycle and differentiate, but rather proliferate continuously and form a tumor cell mass.[24]
It is therefore apparent that mutations in asymmetric cell division are very important for the formation and development of cancer and have significant therapeutic potential.
References
- 1 2 Morrison, S. J.; Kimble, J. (2006). "Asymmetric and symmetric stem-cell divisions in development and cancer". Nature 441 (7097): 1068–1074. doi:10.1038/nature04956. PMID 16810241.
- ↑ Hawkins, Nancy; Garriga, Gian (1998). "Asymmetric cell division: from A to Z.". Genes Dev. 12: 3625–3638. doi:10.1101/gad.12.23.3625.
- ↑ Gönczy, P. and Rose, L.S. Asymmetric cell division and axis formation in the embryo (October 15, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.30.1,
- ↑ Goldstein, B, Hird, SN. "Specification of the anteroposterior axis in Caenorhabditis elegans." Development 1996. 122:1467-74.
- ↑ Cowan, C; Hyman, A (2004). "Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos". Nature 431: 92–6. doi:10.1038/nature02825.
- ↑ O'; Connell, K. Maxwell; White, J. (2000). "The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote". Developmental Biology 222: 55–70. doi:10.1006/dbio.2000.9714.
- ↑ Hamill, DR; Severson, AF; Carter, JC; Bowerman, B (2002). "Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains". Dev. Cell 3: 673–84. doi:10.1016/s1534-5807(02)00327-1.
- ↑ Sadler, PL; Shakes, DC (2000). "Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo". Development 127: 355–66.
- ↑ Cheeks, RJ; Canman, JC; Gabriel, WN; Meyer, N; Strome, S; Goldstein, B (2004). "C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes". Curr Biol 14: 851–62. doi:10.1016/j.cub.2004.05.022.
- ↑ Gönczy, P. and Rose, L.S. Asymmetric cell division and axis formation in the embryo (October 15, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi:10.1895/wormbook.1.30.1
- ↑ Schneider, SQ; Bowerman, B (2003). "Cell polarity and the cytoskeleton in the Caenorhabditis elegans zygote". Annu Rev Genet 37: 221–49.
- 1 2 3 4 5 Matsuzaki, F (2000). "Asymmetric division of Drosophila neural stem cells: a basis for neural diversity". Current Opinion in Neurobiology 10: 38–44. doi:10.1016/s0959-4388(99)00052-5.
- ↑ Guo, M; Jan, LY; Jan, YN (1996). "Control of daughter cell fates during asymmetric division: interaction of Numb and Notch". Neuron 17: 27–41. doi:10.1016/s0896-6273(00)80278-0.
- ↑ Ikeshima-Kataoka, H; Skeath, JB; Nabeshima, Y; Doe, CQ; Matsuzaki, F (1997). "Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions". Nature 390 (6660): 625–629. doi:10.1038/37641. PMID 9403694.
- 1 2 3 4 Henry, Jonathan J.; Martindale, Mark Q. (1999). "Conservation and innovation in spiralian development". Hydrobiologia 402: 255–265. doi:10.1007/978-94-017-2887-4_15.
- 1 2 Shimizu, T.; et al. (1998). "Unequal Cleavage in the early Tubifex Embryo". Develop. Growth Differ 40: 257–266. doi:10.1046/j.1440-169x.1998.00001.x.
- 1 2 Ren, Xiaoyun; Weisblat, David A. (2006). "Asymmetrization of first cleavage by transient disassembly of one spindle pole aster in the leech Helobdella robusta". Developmental Biology 292: 103–115. doi:10.1016/j.ydbio.2005.12.049.
- 1 2 Lambert, J. David; Nagy, Lisa M. (2002). "Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages". Nature 420 (6916): 682–6. doi:10.1038/nature01241. PMID 12478296.
- ↑ Jan, Y. N., and Jan, L. Y. (1998). Nature 392: 775–778. doi:10.1038/33854. Missing or empty
|title=(help) - ↑ Gho, M., Bellaiche, Y., and Schweisguth, F. (1999). "Revisiting the Drosophila microchaete lineage: a novel intrinsically asymmetric cell division generates a glial cell". Development 126: 573–3584.
- ↑ Hayes, A. J., Tudor, D., Nowell, M. A., Caterson, B., and Hughes, C. E (2008). "Chondroitin sulfate sulfation motifs as putative biomarkers for isolation of articular cartilage progenitor cells". J Histochem Cytochem 56: 125–138. doi:10.1369/jhc.7a7320.2007.
- ↑ S. Gómez-López, R. G. Lerner, C. Petritsch, Asymmetric cell division of stem and progenitor cells during homeostasis and cancer. Cell Mol Life Sci, (2013); published online EpubJun (10.1007/s00018-013-1386-1).
- ↑ Xin, H. W.; Ambe, C. M.; Ray, S.; Kim, B. K.; Koizumi, T.; Wiegand, G. W.; Hari, D.; Mullinax, J. E.; Jaiswal, K. R.; Garfield, S. H.; Stojadinovic, A.; Rudloff, U.; Thorgeirsson, S. S.; Avital, I. (2013). "Wnt and the cancer niche: paracrine interactions with gastrointestinal cancer cells undergoing asymmetric cell division". J Cancer 4: 447–457. doi:10.7150/jca.6896.
- ↑ Gonzalez, C. (2007). "Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells". Nat Rev Genet 8: 462–472. doi:10.1038/nrg2103.
Further reading
- Asymmetric Cell Division, Progress in Molecular and Subcellular Biology, volume 45, A. Macieira-Coelho, Editor. Springer Verlag, Berlin, Heidelberg, New York (2007), ISBN 978-3-540-69160-0