Variable surface glycoprotein
Variable surface glycoproteins (VSG) are a type of proteins coating the surface of some infectious microorganisms (e.g. Trypanosoma brucei) and helping them to evade the host's immune system by mean of antigenic variation. The VSG protein is a key molecule for immune escape and parasitic success.
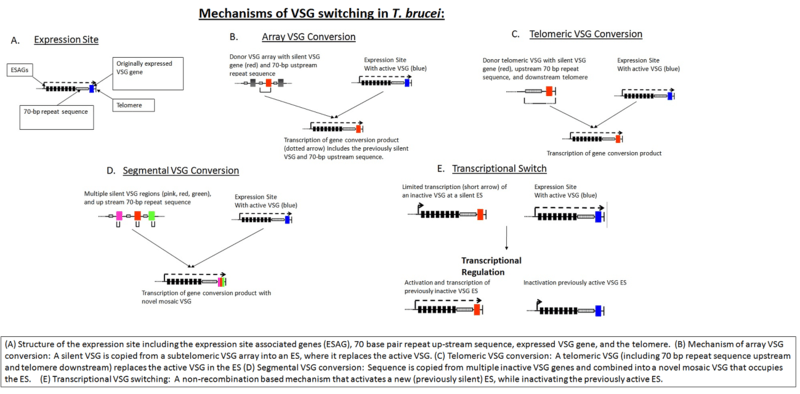
Combinatorial processes increase the diversity of variable surface glycoproteins.[1] The parasite is expressing a series of antigenically distinct VSGs from an estimated 1000 VSG genes. The genes are located in subtelomeric region and are often activated by the duplicative transposition of a silent basic copy gene into an unlinked telomerically located expression site, producing an active expression-linked copy of that gene.[2]
In Trypanosoma brucei
| Variable surface glycoprotein | |
|---|---|
| Identifiers | |
| Organism | |
| Symbol | Tb927.5.4730 |
| Alt. symbols | Tb05.26C7.380 |
| Entrez | 3657576 |
| Other data | |
| Chromosome | 5: 1.41 - 1.41 Mb |
| Variant surface glycoprotein MITAT 1.2 | |
|---|---|
| Identifiers | |
| Organism | |
| Symbol | ? |
| Alt. symbols | VSG 221 |
| UniProt | P26332 |
| Other data | |
In Trypanosoma brucei (see Trypanosoma_brucei#VSG_coat), the variable surface glycoprotein is a major surface antigen.[3]
The cell surface of the bloodstream form features a dense coat of variable surface glycoproteins which is replaced by an equally dense coat of procyclins when the parasite differentiates into the procylic form in the tsetse fly midgut. There is a very fast inhibition of VSG gene transcription which occurs as soon as the temperature is lowered.[4]
The VSGs from T. brucei are attached to the plasma membrane via a glycosyl-phosphatidylinositol (GPI) anchor.[5]
The coat is composed of VSG dimers and forms a macromolecular diffusion barrier. Glycoproteins vary in primary amino acid sequence, the number of N-glycosylation sites, and the types of N-linked oligosaccharides and glycosylphosphatidylinositol membrane anchors they contain. VSG MITat.1.5 is glycosylated at all three potential N-glycosylation sites.[6]
Expression
The bloodstream expression site (BES), or telomeric expression site, is used for changing different variable surface glycoproteins when in host's blood stream to escape the complement system. BESs are polymorphic in size and structure but reveal a surprisingly conserved architecture in the context of extensive recombination. Very small BESs do exist and many functioning BESs do not contain the full complement of expression site associated genes (ESAGs).[7] There is a collection of an estimated 20-30 sites, each being active at a time.[8] Active VSG expression sites are depleted of nucleosomes.[9]
The gene repertoires in T. brucei have diverged to become strain-specific.[10]
The variable surface glycoprotein genes of T. brucei have been classified into two groups depending upon whether or not duplication of the genes is observed when they are expressed.[11]
In other Trypanosoma species
Variable surface glycoproteins are also found in other Trypanosoma species,
In Trypanosoma equiperdum, a parasaite causing the covering sickness in horses, These proteins allow the parasite to efficiently evade the host animal's immune system.[12] These VSGs allow the organism to constantly manipulate and change the surface structure of its proteins, which means it is constantly being presented to the immune system as a new foreign organism and this prevents the body from mounting a large enough immune response to eradicate the disease.[12] In this sense, Trypanosoma equiperdum is a very efficient organism; it may infect less species than other diseases, but it infects and survives very efficiently within its specified hosts. The VSG proteins in T. equiperdum are also phosphorylated.[13]
A VSG gene from Trypanosoma evansi, a parasite that causes a form of surra in animals, has been cloned in Escherichia coli. The expressed protein is immunoreactive with all the sera combinations. The animals immunized with whole cell lysate or recombinant protein show similar antibody reactions in ELISA (Enzyme-linked immunosorbent assay) and CATT (card agglutination test for Trypanosomiasis).[14] The variable surface glycoprotein RoTat 1.2 PCR can be used as a specific diagnostic tool for the detection of T. evansi infections.[15]
The smallest VSG protein (40 kDa in size) to date (1996) has been found in Trypanosoma vivax, which bears little carbohydrate.[16]
In Trypanosoma congolense, in vitro analyses of the incorporated sugars after hydrolysis of the glycoprotein showed that glucosamine and mannose are utilized in the biosynthesis of the carbohydrate moiety directly whereas galactose was converted possibly to other intermediates before being incorporated into the antigen. The unglycosylated VSG with a molecular weight of 47 kDa had completely lost its size heterogeneity.[17]
See also
- Coat protein (disambiguation)
- Glycocalyx
- List of MeSH codes (D23)
- List of MeSH codes (D12.776.395)
- List of MeSH codes (D12.776.543)
- Amastin, another surface (trans-membrane) glycoprotein in trypanosomatid parasites[18]
References
- ↑ Thon G, Baltz T, Giroud C, Eisen H (August 1990). "Trypanosome variable surface glycoproteins: composite genes and order of expression". Genes Dev. 4 (8): 1374–83. doi:10.1101/gad.4.8.1374. PMID 2227415.
- ↑ Buck GA, Jacquemot C, Baltz T, Eisen H (December 1984). "Re-expression of an inactivated variable surface glycoprotein gene in Trypanosoma equiperdum". Gene 32 (3): 329–36. doi:10.1016/0378-1119(84)90008-8. PMID 6530143.
- ↑ Ferguson MA, Homans SW (September 1988). "Parasite glycoconjugates: towards the exploitation of their structure". Parasite Immunol. 10 (5): 465–79. doi:10.1111/j.1365-3024.1988.tb00236.x. PMID 3057422.
- ↑ Pays E, Coquelet H, Pays A, Tebabi P, Steinert M (September 1989). "Trypanosoma brucei: posttranscriptional control of the variable surface glycoprotein gene expression site". Mol. Cell. Biol. 9 (9): 4018–21. PMC 362464. PMID 2779574.
- ↑ D.J. Grab DJ, Verjee Y. "Localization of a Variable Surface Glycoprotein Phosphatidylinositol-Specific Phospholipase-C in Trypanosoma brucei brucei". FAO Corporate document depository. Food and Agricultural Organization of the United Nations.
- ↑ Mehlert A, Bond CS, Ferguson MA (October 2002). "The glycoforms of a Trypanosoma brucei variant surface glycoprotein and molecular modeling of a glycosylated surface coat". Glycobiology 12 (10): 607–12. doi:10.1093/glycob/cwf079. PMID 12244073.
- ↑ Hertz-Fowler C, Figueiredo LM, Quail MA, Becker M, Jackson A, Bason N, Brooks K, Churcher C, Fahkro S, Goodhead I, Heath P, Kartvelishvili M, Mungall K, Harris D, Hauser H, Sanders M, Saunders D, Seeger K, Sharp S, Taylor JE, Walker D, White B, Young R, Cross GA, Rudenko G, Barry JD, Louis EJ, Berriman M (2008). "Telomeric expression sites are highly conserved in Trypanosoma brucei". PLoS ONE 3 (10): e3527. Bibcode:2008PLoSO...3.3527H. doi:10.1371/journal.pone.0003527. PMC 2567434. PMID 18953401.
- ↑ Vanhamme L, Lecordier L, Pays E (May 2001). "Control and function of the bloodstream variant surface glycoprotein expression sites in Trypanosoma brucei". Int. J. Parasitol. 31 (5-6): 523–31. doi:10.1016/S0020-7519(01)00143-6. PMID 11334937.
- ↑ Stanne TM, Rudenko G (January 2010). "Active VSG expression sites in Trypanosoma brucei are depleted of nucleosomes". Eukaryotic Cell 9 (1): 136–47. doi:10.1128/EC.00281-09. PMC 2805301. PMID 19915073.
- ↑ Hutchinson OC, Picozzi K, Jones NG, Mott H, Sharma R, Welburn SC, Carrington M (2007). "Variant Surface Glycoprotein gene repertoires in Trypanosoma brucei have diverged to become strain-specific". BMC Genomics 8: 234. doi:10.1186/1471-2164-8-234. PMC 1934917. PMID 17629915.
- ↑ Young JR, Turner MJ, Williams RO (1984). "The role of duplication in the expression of a variable surface glycoprotein gene of Trypanosoma brucei". J. Cell. Biochem. 24 (3): 287–95. doi:10.1002/jcb.240240309. PMID 6736139.
- ↑ 12.0 12.1 Raibaud A, Gaillard C, Longacre S, Hibner U, Buck G, Bernardi G, Eisen H (July 1983). "Genomic environment of variant surface antigen genes of Trypanosoma equiperdum". Proc. Natl. Acad. Sci. U.S.A. 80 (14): 4306–10. Bibcode:1983PNAS...80.4306R. doi:10.1073/pnas.80.14.4306. PMC 384026. PMID 6308614.
- ↑ Baltz T, Giroud C, Baltz D, Duvillier G, Degand P, Demaille J, Pautrizel R (1982). "The variable surface glycoproteins of Trypanosoma equiperdum are phosphorylated". EMBO J. 1 (11): 1393–8. PMC 553222. PMID 6821334.
- ↑ Sengupta PP, Balumahendiran M, Balamurugan V, Rudramurthy GR, Prabhudas K (June 2012). "Expressed truncated N-terminal variable surface glycoprotein (VSG) of Trypanosoma evansi in E. coli exhibits immuno-reactivity". Vet. Parasitol. 187 (1–2): 1–8. doi:10.1016/j.vetpar.2012.01.012. PMID 22277627.
- ↑ Claes F, Radwanska M, Urakawa T, Majiwa PA, Goddeeris B, Büscher P (2004). "Variable Surface Glycoprotein RoTat 1.2 PCR as a specific diagnostic tool for the detection of Trypanosoma evansi infections". Kinetoplastid Biol Dis 3 (1): 3. doi:10.1186/1475-9292-3-3. PMC 521498. PMID 15377385.
- ↑ Gardiner PR, Nene V, Barry MM, Thatthi R, Burleigh B, Clarke MW (November 1996). "Characterization of a small variable surface glycoprotein from Trypanosoma vivax". Mol. Biochem. Parasitol. 82 (1): 1–11. doi:10.1016/0166-6851(96)02687-4. PMID 8943146.
- ↑ Reinwald E, Heidrich C, Risse HJ (May 1984). "In vitro studies on the biosynthesis of the surface glycoprotein of Trypanosoma congolense". J. Protozool. 31 (2): 300–6. doi:10.1111/j.1550-7408.1984.tb02966.x. PMID 6470988.
- ↑ Jackson AP (January 2010). "The evolution of amastin surface glycoproteins in trypanosomatid parasites". Mol. Biol. Evol. 27 (1): 33–45. doi:10.1093/molbev/msp214. PMC 2794310. PMID 19748930.
External links
- Variant Surface Glycoproteins, Trypanosoma at the US National Library of Medicine Medical Subject Headings (MeSH)
- www.icp.ucl.ac.be